Overview
The laboratory's primary focus is the development of novel computational approaches for computational systems biology and systems pharmacology and their application to identify and understand the biological mechanisms underlying complex diseases and their treatment. We are particularly interested in the development and real-world application of novel computational methods for mathematical modeling in precision medicine to extend the field's capabilities in defining hybrid models of complex diseases potentially integrated with machine learning and AI-based tools. The laboratory is also interested in novel algorithms for biomarker identification, both single and multi-omics.
Research directions
- Multiscale hybrid modeling integrated with machine learning and AI: a challenging modeling aspect is a need to provide a systems-level understanding of the disease by encoding in a unified mathematical description a wide variety of independent or interacting genetic and non-genetic factors acting at different levels of biology (genetic, molecular, tissue, organ, etc.) which concur to the development of a complex disease. To this aim, models can be developed as hierarchical, multiscale, mathematical entities, where the disease is described according to different levels of detail. We design and implement novel hybrid modeling approaches, where the hierarchical layers of the model are potentially developed with different computational techniques (boolean networks, discrete models, ordinary differential equations, machine learning, AI-based tools, etc.) and then integrated to form a unique model able to better adapt to the amount of biological knowledge and data that is available for each hierarchical layer.
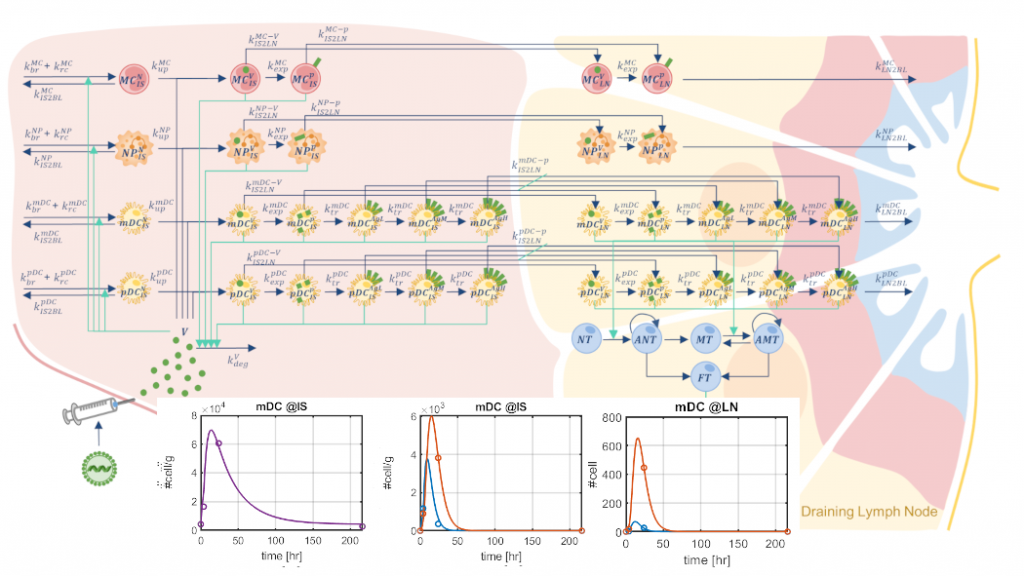
- Real-world modeling applications for systems pharmacology and precision medicine: in addition to developing modeling techniques, we also develop novel computational algorithms to apply mathematical modeling to highly challenging applications in systems pharmacology and precision medicine. We promote the development of models of complex diseases in several research areas (e.g., cancer, metabolism, neurodegeneration, infectious diseases, etc.). We design algorithms that exploit the generated models to analyze the individual variability of a patient population, predict treatment efficacy, and identify novel treatment protocols (e.g., algorithms for model-based patient stratification, in silico generation of virtual populations).
- Hybrid stochastic simulation and sensitivity analysis: once a mathematical model is defined, it can be analyzed by simulation and sensitivity analysis to predict the behavior in time of the system and identify the leading regulative players. Stochastic algorithms provide an accurate way of simulating and analyzing a biological system since they can reproduce their random and discrete nature. Deterministic algorithms provide faster solutions, but results are approximated, and stochasticity is not considered. We promote the development of hybrid simulation and sensitivity analysis methods to combine exact and approximate algorithms in the same computational process. The speed-up of the computation provided by the developed hybrid methods allows to analyze larger models by preserving the accuracy of the computed results as much as possible. This could open new perspectives in computational systems biology, where scientists often have to find a compromise between the accuracy of their simulations and the need to consider vast and complex reaction networks.
- Algorithms for multi-omics biomarker identification: thanks to the exponential development in high-throughput technologies, multiple layers of biological data are becoming available for the characterization of disease mechanisms on a large scale. Therefore, effective integration of omics data is needed to gain maximal extraction of biological information and identify reliable biomarkers. We promote the development and implementation of novel computational methods based on meta-dimensional and multi-stage approaches for integrating omics data encompassing genetics, RNA, and protein expression on a genome-scale. Such methods will be applied to identify disease biomarkers in several research areas, such as cancer, metabolic diseases, and neurodegenerative disorders.
Group members
- Luca Marchetti, PI
- Giada Fiandaca, postdoctoral fellow
- Natascia Zangani, postdoctoral fellow
- Lorenzo Dasti, postdoctoral fellow
- Giuseppe Fiorentino, Ph.D. student, Doctoral Program in Biomolecular Sciences, Department CIBIO
- Anna Fochesato, Ph.D. student, Department of Mathematics
- Emiliano Maresi, Ph.D. student, Department of Mathematics
- Elena Righetti, Ph.D. student, Doctoral Program in Biomolecular Sciences, Department CIBIO
- Elio Campanile, Ph.D. student, Department of Mathematics
- Stefano Giampiccolo, Ph.D. student, Doctorate Programme in Industrial Innovation
Grants and Contract Research
2021, Wellcome Leap Program “R3: RNA Readiness and Response”
Collaborations
- Fondazione The Microsoft Research – University of Trento Centre for Computational and Systems Biology (COSBI), Rovereto, Italy
- European Institute of Oncology (IEO), Milan, Italy
- Cornell University, USA
- Texas A&M University, USA
- Emilio Siena, Data Science and Computational Vaccinology, GSK, Siena, Italy
- Riccardo Bonadonna, Department of Medicine, University of Parma, Italy
- Andrea Pugliese, Department of Mathematics, University of Trento, Italy
- Orazio Caffo, Medical Oncology Department - Santa Chiara Hospital, Trento, Italy
- Damiano Bragantini, Infectious Diseases Unit, Pederzoli Hospital, Peschiera del Garda, Italy
- Pierluigi Strippoli and Allison Piovesan, University of Bologna, Italy
- Lucia Carboni, Department of Pharmacy and Biotechnology, University of Bologna, Italy
Funding
Bando: PRIN 2022 (D.D. 104/22)
PREDICTive Cardio-metabolic transcriptOMIC trajectorieS in the Barilla Offspring Follow-up STUDY: The PREDICT-OMICS Study
Luca Marchetti, Responsabile di Unità
Codice Protocollo: 2022FZL247 CUP: E53D23013170001

Selected publications
* indicates equally contributing authors.
A. Paris, P. Bora, S. Parolo, D. MacCannell, M. Monine, N. van der Munnik, X. Tong, S. Eraly, Z. Berger, D. Graham, T. Ferguson, E. Domenici, I. Nestorov, L. Marchetti, “A pediatric quantitative systems pharmacology model of neurofilament trafficking in spinal muscular atrophy treated with the antisense oligonucleotide nusinersen”. CPT Pharmacometrics and Systems Pharmacology 12:196-206, 2023.
E. Piretto, G. Selvaggio, D. Bragantini, E. Domenici, L. Marchetti, “A novel logical model of COVID-19 intracellular infection to support therapies development”. PLoS Computional Biology 18(8): e1010443, 2023.
A. Paris, P. Bora, S. Parolo, M. Monine, X. Tong, S. Eraly, E. Masson, T. Ferguson, A. McCampbell, D. Graham, E. Domenici, I. Nestorov, L Marchetti, “An age-dependent mathematical model of neurofilament trafficking in healthy conditions”. CPT Pharmacometrics and Systems Pharmacology, 11:447-457, 2022.
R. Coletti, A. Pugliese, A. Lunardi, O. Caffo, L. Marchetti, “A Model-Based Framework to Identify Optimal Administration Protocols for Immunotherapies in Castration-Resistance Prostate Cancer”. Cancers 2022, 14, 135.
G. Selvaggio, L. Leonardelli, G. Lofano, S. Fresnay, S. Parolo, D. Medini, E. Siena*, L. Marchetti*, “A quantitative systems pharmacology approach to support mRNA vaccine development and optimization”. CPT Pharmacometrics and Systems Pharmacology, 10:1448-1451, 2021.
L. Leonardelli*, G. Lofano*, G. Selvaggio, S. Parolo, S. Giampiccolo, D. Tomasoni, E. Domenici, C. Priami, H. Song, D. Medini, L. Marchetti*, E. Siena*, “Literature Mining and Mechanistic Graphical Modelling to Improve mRNA Vaccine Platforms”. Frontiers in Immunology, 12, 738388, 2021.
G. Fiorentino, R. Visintainer, E. Domenici, M. Lauria, L. Marchetti, “MOUSSE: Multi-Omics Using Subject-Specific SignaturEs”. Cancers 2021, 13, 3423.
A. Fochesato*, G. Simoni*, F. Reali, G. Giordano, E. Domenici, L. Marchetti, “A Retrospective Analysis of the COVID-19 Pandemic Evolution in Italy”. Biology 10:311, 2021.
R. Coletti, A. Pugliese, L. Marchetti, “Modeling the effect of immunotherapies on human castration-resistant prostate cancer”. Journal of Theoretical Biology 509, 110500, 2021.
G. Simoni, C. Kaddi, M.Tao, F. Reali, D. Tomasoni, C. Priami, K. Azer, S. Neves-Zaph, L. Marchetti, “A robust computational pipeline for model-based and data-driven phenotype clustering''. Bioinformatics 37(9):1269-1277, 2020.
R. Coletti, L. Leonardelli, S. Parolo, L. Marchetti, “A QSP model of prostate cancer immunotherapy to identify effective combination therapies''. Scientific Reports 10, Article number: 9063, 2020.
L. Marchetti, M. Lauria, L. Caberlotto, L. Musazzi, M. Popoli, A. A. Mathée, E. Domenici, L. Carboni, “Gene expression signature of antidepressant treatment response/non-response in Flinders Sensitive Line rats subjected to maternal separation''. European Neuropsychopharmacology 31:69-85, 2020.
G. Simoni, H. T. Vo, C. Priami, L. Marchetti, “A comparison of deterministic and stochastic approaches for sensitivity analysis in computational systems biology''. Briefings in Bioinformatics, 21(2):527-540, 2019.
K. Misselbeck*, L. Marchetti*, C. Priami, P. J. Stover*, M. S. Field*, “The 5-formyltetrahydrofolate futile cycle reduces pathway stochasticity in an extended hybrid-stochastic model of folate-mediated one-carbon metabolism" Scientific Reports 9, Article number: 4322, 2019.
C. Uluseker*, G. Simoni*, L. Marchetti, M. Dauriz, A. Matone, C. Priami, “A closed-loop multi-level model of glucose homeostasis''. PLoS ONE 13(2):e0190627, 2018.
L. Marchetti, C. Priami, Vo H. T., “Simulation Algorithms for Computational Systems Biology". Springer International Publishing. ISBN: 978-3-319-63113-4, 2017.
G. Tini, L. Marchetti, C. Priami, M. P. Scott-Boyer, “Multi-omics integration—a comparison of unsupervised clustering methodologies''. Briefings in Bioinformatics 20(4), 1269-1279, 2017.
L. Marchetti*, E. Siena*, M. Lauria, D. Maffione, N. Pacchiani, C. Priami, D. Medini, “Exploring the limitations of peripheral blood transcriptional biomarkers in predicting influenza vaccine responsiveness”. Complexity, Volume 2017, Article ID 3017632, 2017.
L. Marchetti*, F. Reali*, M. Dauriz, C. Brangani, L. Boselli, G. Ceradini, E. Bonora, R. C. Bonadonna, C. Priami, “A Novel Insulin/Glucose Model after a Mixed-Meal Test in Patients with Type 1 Diabetes on Insulin Pump Therapy". Scientific Reports 6, Article number: 36029, 2016.
L. Marchetti, C. Priami, Vo H. T., “HRSSA – Efficient hybrid stochastic simulation for spatially homogeneous biochemical reaction networks". Journal of Computational Physics, Vol. 317, pag. 301-317, 2016.
F. Capuani*, A. Conte*, E. Argenzio, L. Marchetti, C. Priami, S. Polo, P. P. Di Fiore*, S. Sigismund*, A. Ciliberto*, “Quantitative analysis reveals how EGFR activation and downregulation are coupled in normal but not in cancer cells". Nature Communications, Vol. 6, No. 7999, 2015.
For a complete list see: LINK
