Overview
Human diseases such as cancer are intrinsically entangled with complexity. The discovery of effective cures requires dealing with this complexity and greatly benefits from the development of high-resolution methods of investigation: genome-wide, multi-modal, single cell, spatially resolved. Understanding human diseases at single-cell resolution within their architectural context is a scientific challenge requiring dedicated computational analysis dealing with the volume and heterogeneity of the data. The research mission of the lab is to understand the RNA molecular mechanisms underlying dysregulation in human diseases, by combining high-throughput and high-resolution analyses, with a pan-disciplinary approach. 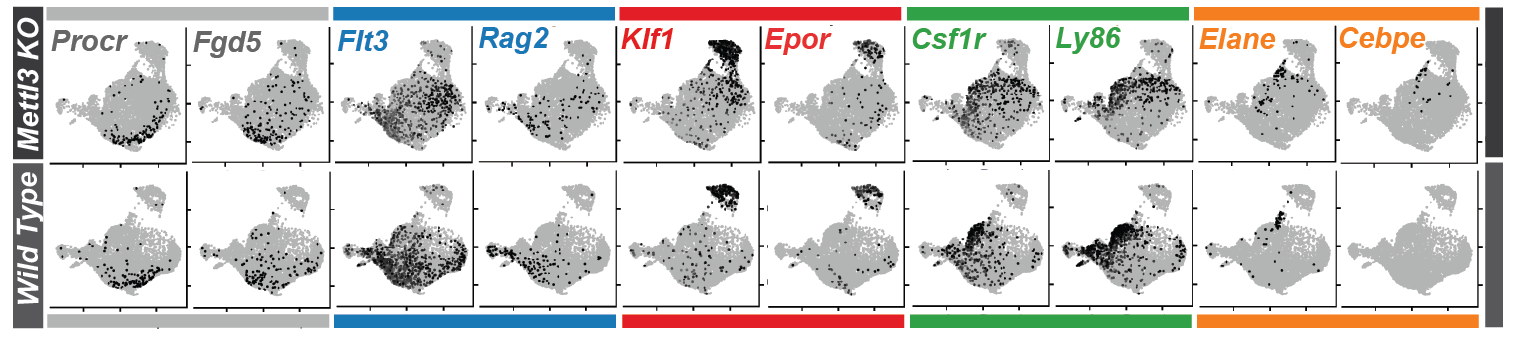
Research directions
-
Study human disease by single-cell and spatial omics
Thanks to the revolution of single cell sequencing, today we can obtain sequencing data from single cells. By looking at thousands of cells one at a time, we can see which set of genes each individual cell is transcribing, and we can capture the cellular diversity of tissues with unprecedented resolution. Single cell and spatially resolved data analysis requires the parallel development of appropriate methods for the identification of cell types, gene regulatory networks, spatial expression patterns and dependencies. The research of the lab is focused on multi-modal analysis of single cell and spatially resolved data to understand the molecular architecture of gene expression in human diseases.
-
Develop computational strategies for RNA data science, multi-omics integration and visualization.
The lab develops computational methods for the high-resolution analysis, integration and visualization of multi-omics data: RNA and protein levels, RNA splicing, structure and modifications, RNA-protein interactions, translation. The availability of public big data sets such as the Expression Atlas, ENCODE and the Human Cell Atlas also provides unprecedented opportunities and challenges for data mining, deep analysis and knowledge extraction from various omics layers concerning RNA biology. -
Chart RNA modifications, structures and interactions in health and disease.
Coding and non-coding RNA molecules are post-transcriptionally modified and dynamically interact with RNA-binding proteins and other RNAs to form ribonucleoprotein complexes (RNPs) and membrane-less organelles. This complex network of modifications and interactions ultimately regulate the existence and function of RNAs within cells. Physiological RNA modifications and interactions are frequently altered in human pathologies such as neurodegenerative diseases and tumors. To understand how mutations affect RNA-protein complexes, the lab integrates next-generation sequencing techniques (e.g. CLIP-Seq) and computational biology to obtain comprehensive maps of protein-RNA, RNA-RNA and RNA-DNA interactions with high resolution. -
Follow translation dynamics
Cells regulate protein synthesis by tuning the translation of specific genes. Translation affects protein levels as much as transcription but with mechanisms that are less studied. The last few years have witnessed a rapid adoption of ribosome profiling as a powerful technique to study translation at the genome-wide level and providing unique information on ribosome positions along RNAs. The lab uses ribosome profiling and proteomics data to monitor translation regulation dynamics in health and disease.
Group members
- Toma Tebaldi, PI
- Emma Busarello, PhD student
- Christian Ramirez Amarilla, PhD student
- Gabriele Tomè, Post-graduate QCB (with Dompè farmaceutici)
- Irene Moratti, Master student QCB
- Zehra Korkusuz, Master student DISI
Bachelor and Master Degree Thesis projects are currently available. The laboratory is looking for PhD and Post Doctoral candidates.
If interested, please contact toma.tebaldi [at] unitn.it
Collaborations
- Stephanie Halene (Yale University, US)
- Diane Krause (Yale University, US)
- Emanuela Bruscia (Yale University, US)
- Rong Fan (Yale University, US)
- Anna Mandinova (Harvard Medical School, US)
- Therese Standal (NTNU, Norway)
- Gabriella Viero (Institute of Biophysics, CNR, Italy)
- Enrico Blanzieri (DISI - University of Trento, Italy)
- Alessandro Fatica (University of Roma - Sapienza, Italy)
- Piergiorgio Amendola (Dompè Farmaceutici s.p.a)
Funding
- 2021-2025 - My First AIRC Grant
Multimodal single-cell analysis of hematologic malignancies
Selected publications
Biancon G, Busarello E, Joshi P, Lesch BJ, Halene S, Tebaldi T#. Deconvolution of in vivo protein-RNA contacts using fractionated eCLIP-seq. STAR Protoc. 2022 Dec 16;3(4):101823. (#Co-corresponding autorship)
Antonica F, Santomaso L, Pernici D, Petrucci L, Aiello G, Cutarelli A, Conti L, Romanel A, Miele E, Tebaldi T, Tiberi L. A slow-cycling/quiescent cells subpopulation is involved in glioma invasiveness. Nat Commun. 2022 Aug 15;13(1):4767.
Biancon G, Joshi P, Zimmer JT, Hunck T, Gao Y, Lessard MD, Courchaine E, Barentine AES, Machyna M, Botti V, Qin A, Gbyli R, Patel A, Song Y, Kiefer L, Viero G, Neuenkirchen N, Lin H, Bewersdorf J, Simon MD, Neugebauer KM, Tebaldi T#^, Halene S. Precision analysis of mutant U2AF1 activity reveals deployment of stress granules in myeloid malignancies. Mol Cell. 2022 Mar 17;82(6):1107-1122.e7. (#Co-corresponding authorship, ^Journal cover)
Gao Y, Chen S, Halene S, Tebaldi T#. Transcriptome-wide quantification of double-stranded RNAs in live mouse tissues by dsRIP-Seq. STAR Protoc. 2021 Mar 18;2(1):100366. (#Co-corresponding autorship)
Song Y, Shan L, Gbyli R, Liu W, Strowig T, Patel A, Fu X, Wang X, Xu ML, Gao Y, Qin A, Bruscia EM, Tebaldi T, Biancon G, Mamillapalli P, Urbonas D, Eynon E, Gonzalez DG, Chen J, Krause DS, Alderman J, Halene S, Flavell RA. Combined liver-cytokine humanization comes to the rescue of circulating human red blood cells. Science. 2021 Mar 5;371(6533):1019-1025.
Liu Y, Yang M, Deng Y, Su G, Enninful A, Guo CC, Tebaldi T, Zhang D, Kim D, Bai Z, Norris E, Pan A, Li J, Xiao Y, Halene S, Fan R. High-Spatial-Resolution Multi-Omics Sequencing via Deterministic Barcoding in Tissue. Cell. 2020 Dec 10;183(6):1665-1681.e18.
Gao Y, Vasic R, Song Y, Teng R, Liu C, Gbyli R, Biancon G, Nelakanti R, Lobben K, Kudo E, Liu W, Ardasheva A, Fu X, Wang X, Joshi P, Lee V, Dura B, Viero G, Iwasaki A, Fan R, Xiao A, Flavell RA, Li HB, Tebaldi T#, Halene S. m6A Modification Prevents Formation of Endogenous Double-Stranded RNAs and Deleterious Innate Immune Responses during Hematopoietic Development. Immunity. 2020 Jun 16;52(6):1007-1021.e8. (#Co-corresponding autorship)
Lauria F*, Bernabò P*, Tebaldi T*, Groen EJN*, Perenthaler E, Maniscalco F, Rossi A, Donzel D, Clamer M, Marchioretto M, Omersa N, Orri J, Dalla Serra M, Anderluh G, Quattrone A, Inga A, Gillingwater TH, Viero G. SMN-primed ribosomes modulate the translation of transcripts related to spinal muscular atrophy. Nat Cell Biol. 2020 Oct;22(10):1239-1251. (*Co-first autorship)
Zubovic L, Piazza S, Tebaldi T, Cozzuto L, Palazzo G, Sidarovich V, De Sanctis V, Bertorelli R, Lammens T, Hofmans M, De Moerloose B, Ponomarenko J, Pigazzi M, Masetti R, Mecucci C, Basso G, Macchi P. The altered transcriptome of pediatric myelodysplastic syndrome revealed by RNA sequencing. J Hematol Oncol. 2020 Oct 12;13(1):135.
Murthy V*, Tebaldi T*, Yoshida T, Erdin S, Calzonetti T, Vijayvargia R, Tripathi T, Kerschbamer E, Seong IS, Quattrone A, Talkowski ME, Gusella JF, Georgopoulos K, MacDonald ME, Biagioli M. Hypomorphic mutation of the mouse Huntington’s disease gene orthologue. PLoS Genet. 2019 Mar 21;15(3):e1007765 (*Co-First autorship)
Lauria F*, Tebaldi T*, Bernabò P, Groen EJN, Gillingwater TH, Viero G. riboWaltz: Optimization of ribosome P-site positioning in ribosome profiling data. PLoS Comput Biol. 2018 Aug 13;14(8):e1006169. (*Co-First autorship)
Tebaldi T*^, Zuccotti P*, Peroni D*, Köhn M*, Gasperini L, Potrich V, Bonazza V, Dudnakova T, Rossi A, Sanguinetti G, Conti L, Macchi P, D’Agostino V, Viero G, Tollervey D, Hüttelmaier S, Quattrone A. HuD is a neural translation enhancer acting on mTORC1-responsive genes and counteracted by the Y3 small non-coding RNA. Mol Cell. 2018 Jul 19;71(2):256-270.e10. (*Co-First autorship, ^With a dedicated Journal Preview: "A Translation Tuning HuDdle for Neurons")
Liang Y*, Tebaldi T*^, Rejeski K*, Joshi P*, Stefani G*, Taylor A, Song Y, Vasic R, Maziarz J, Balasubramanian K, Ardasheva A, Ding A, Quattrone A, Halene S. SRSF2 mutations drive oncogenesis by activating a global program of aberrant alternative splicing in hematopoietic cells. Leukemia. 2018 Jun 1 (*Co-First autorship, ^Recommended in F1000Prime as being of special significance in its field)
Negro S*, Stazi M*, Marchioretto M*, Tebaldi T*, Rodella U, Duregotti E, Gerke V, Quattrone A, Montecucco C, Rigoni M, Viero G. Hydrogen peroxide is a neuronal alarmin that triggers specific RNAs, local translation of Annexin A2 and cytoskeletal remodeling in Schwann cells. RNA. 2018 Jul;24(7):915-925 (*Co-First autorship)
Clamer M, Tebaldi T^, Lauria F, Bernabò P, Gómez-Biagi RF, Marchioretto M, Kandala DT, Minati L, Perenthaler E, Gubert D, Pasquardini L, Guella G, Groen EJN, Gillingwater TH, Quattrone A, Viero G. Active Ribosome Profiling with RiboLace. Cell Rep. 2018 Oct 23;25(4):1097-1108.e5. (^Journal cover)
Bernabò P*, Tebaldi T*^, Groen EJN*, Lane FM*, Perenthaler E, Mattedi F, Newbery HJ, Zhou H, Zuccotti P, Potrich V, Shorrock HK, Muntoni F, Quattrone A, Gillingwater TH, Viero G. In Vivo Translatome Profiling in Spinal Muscular Atrophy Reveals a Role for SMN Protein in Ribosome Biology. Cell Rep. 2017 Oct 24;21(4):953-965. (*Co-First authorship, ^Journal cover)
Corrado G, Tebaldi T, Costa F, Frasconi P, Passerini A. RNAcommender: genome-wide recommendation of RNA-protein interactions. Bioinformatics. 2016 Dec 1;32(23):3627-3634.
Tebaldi T*#, Zaccara S*, Alessandrini F, Bisio A, Ciribilli Y, Inga A#. Whole-genome cartography of p53 response elements ranked on transactivation potential. BMC Genomics. 2015 Jun 17;16:464. (*Co-First authorship, #Co-Corresponding autorship)
Tebaldi T*, Re A*, Viero G, Pegoretti I, Passerini A, Blanzieri E, Quattrone A. Widespread uncoupling between transcriptome and translatome variations after a stimulus in mammalian cells. BMC Genomics. 2012 Jun 6;13:220. (*Co-First autorship)
Complete List of Published Work available here.
