Overview
 Main focus of the laboratory is to determine the crystallographic structures of proteins or protein complexes with a pathogenetic role in various human diseases. This knowledge is then exploited to develop chemical probes to better understand their functions and regulation mechanisms and in producing hit and lead compounds of potential pharmacological interest.
Main focus of the laboratory is to determine the crystallographic structures of proteins or protein complexes with a pathogenetic role in various human diseases. This knowledge is then exploited to develop chemical probes to better understand their functions and regulation mechanisms and in producing hit and lead compounds of potential pharmacological interest.
Research directions
-
Bromodomains
Bromodomains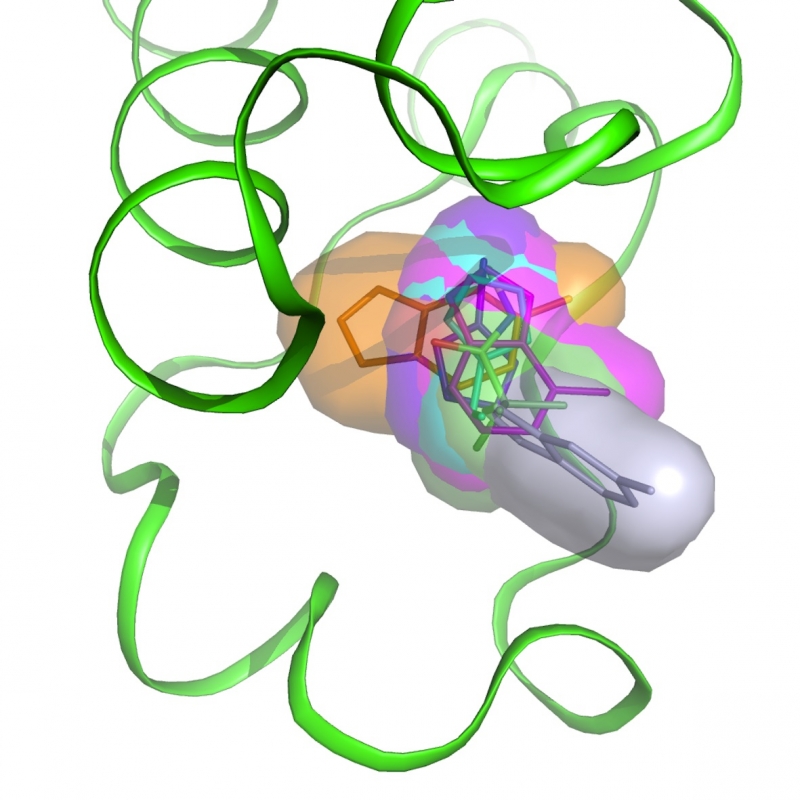 are epigenetic reader modules recognizing acetylated lysines in histones and other proteins, regulating transcription and involved in various malignancies. The members of the BET subfamily of human bromodomains have been extensively studied in recent years with 12 BET inhibitors currently tested in clinical trials. Less is known about the function of other bromodomain-containing proteins such as BAZ2A and BAZ2B. As for BET bromodomains, the development of potent and selective inhibitors suitable for phenotypic profiling would help uncovering their biological roles. These could also prove therapeutically beneficial.
are epigenetic reader modules recognizing acetylated lysines in histones and other proteins, regulating transcription and involved in various malignancies. The members of the BET subfamily of human bromodomains have been extensively studied in recent years with 12 BET inhibitors currently tested in clinical trials. Less is known about the function of other bromodomain-containing proteins such as BAZ2A and BAZ2B. As for BET bromodomains, the development of potent and selective inhibitors suitable for phenotypic profiling would help uncovering their biological roles. These could also prove therapeutically beneficial. -
YTH domains
YTH domains are epitranscriptomic reader modules recognising methyladenosine-containing RNAs and regulating mRNA stability. Inhibitors of these domains would help in defining their involvement in various physiological and pathological pathways. -
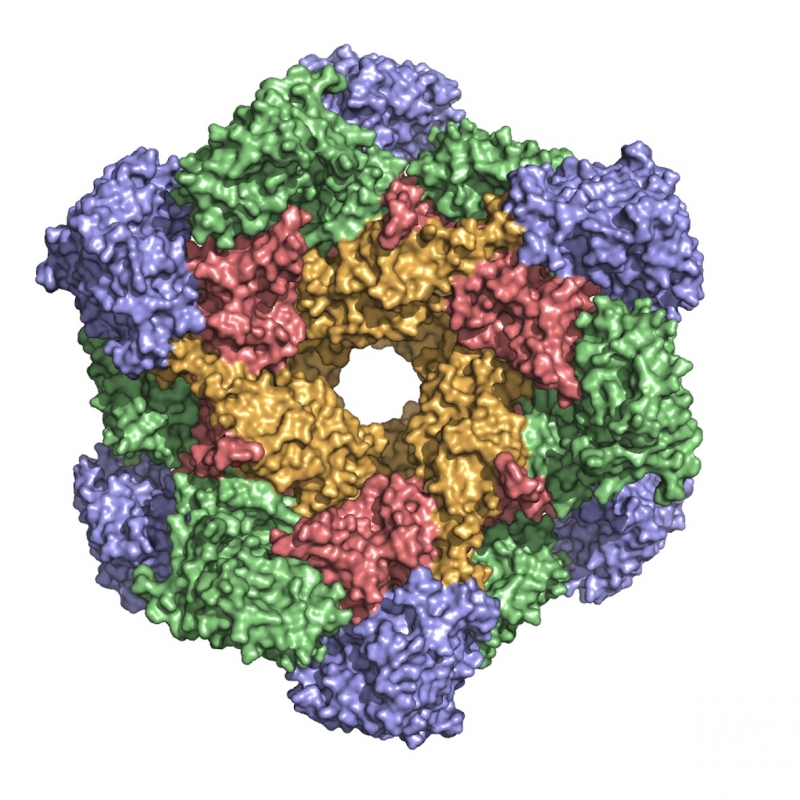
CK2
CK2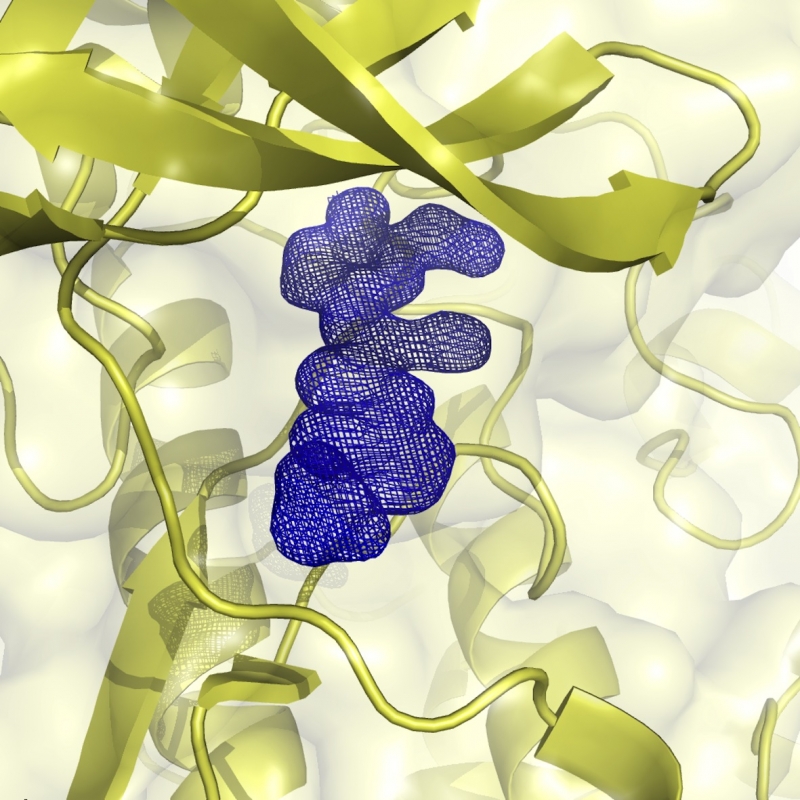 is a protein kinase involved in diverse cellular processes from apoptosis to differentiation. Its activity has been found elevated in various cancers, especially haematological malignancies. Its anti-apoptotic role has been associated with its involvement in sustaining cancer growth and in protecting cancerous cells. A number of small molecules interfering with CK2 activity have been developed over the last years culminating in the so far unique CK2 inhibitor in clinical trials, CX-4945. The laboratory is actively involved in defining CK2 unusual self-inhibitory regulation mechanism and in developing new molecules of potential pharmacological interest as inhibitors of the human kinase (cancer) and of parasitic kinases (malaria, leishmaniasis and trypanosomiasis).
is a protein kinase involved in diverse cellular processes from apoptosis to differentiation. Its activity has been found elevated in various cancers, especially haematological malignancies. Its anti-apoptotic role has been associated with its involvement in sustaining cancer growth and in protecting cancerous cells. A number of small molecules interfering with CK2 activity have been developed over the last years culminating in the so far unique CK2 inhibitor in clinical trials, CX-4945. The laboratory is actively involved in defining CK2 unusual self-inhibitory regulation mechanism and in developing new molecules of potential pharmacological interest as inhibitors of the human kinase (cancer) and of parasitic kinases (malaria, leishmaniasis and trypanosomiasis).
-
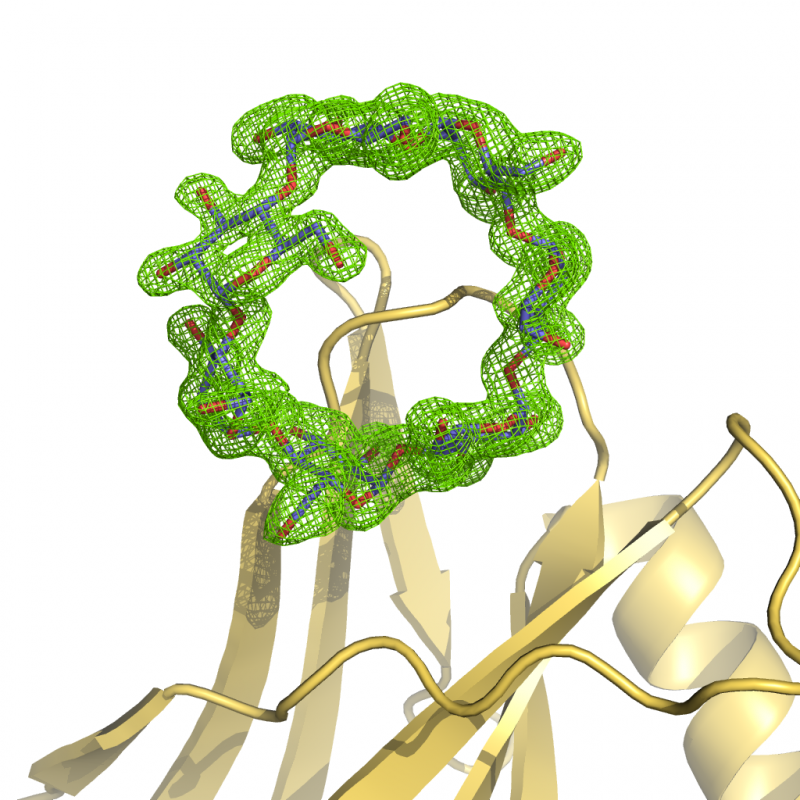
Lafora Disease
Lafora disease (LD) is a teenage-onset pathology characterized by spasmodic jerky contractions and epileptic seizures, which become increasingly frequent in the next years. Hallucination, dementia and vegetative state follow. Death occurs around age 25. The hallmark of LD is the presence in neurons of large neurotoxic inclusions, composed of aberrant and insoluble glycogen molecules. Structural dissection of the protein complexes involved in LD development and prognosis would help in identifying possible points of pharmacological intervention to subsequently target with small molecule inhibitors.
Group members
- Graziano Lolli, PI
- Giulia Cazzanelli, postdoctoral fellow
- Andrea Dalle Vedove, postdoctoral fellow
- Gabriele Trentini, PhD student
- Eleonora Parolin, PhD student
Funding
- Alzheimer’s Association NTF grant 2022-2024
- My First AIRC Grant 2018-2022
-
Bando: PRIN 2022 (D.D. 104/22)
Therapeutic opportunities in Lafora disease, a rare juvenile neurodegenerative epilepsy with no treatment
Graziano Lolli, Coordinatore
Codice Protocollo: 2022WHPWAL CUP: E53D23010050006
Collaborations
- Prof. Amedeo Caflisch, University of Zurich, Switzerland
- Prof. Alessandro Quattrone, Alessandro Provenzani, Vito Giuseppe D’Agostino, Luca Fava, Ines Mancini, University of Trento, Italy
- Dr. Paola Storici, Elettra Synchrotron, Trieste, Italy
- Dr. Orazio Palumbo, Fondazione IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo, Italy
- Dr. Giuseppe d’Orsi, Ospedali Riuniti and University of Foggia, Italy
- Prof. Maria Letizia Barreca, University of Perugia, Italy
- Dr. Gabriele Giachin, University of Padua, Italy
Selected publications
Dalle Vedove A., Cazzanelli G., Corsi J., Sedykh M., D’Agostino V.G., Caflisch A. and Lolli G. (2021) Identification of a BAZ2A bromodomain hit compound by fragment joining. ACS Bio & Med Chem Au. https://doi.org/10.1021/acsbiomedchemau.1c00016
Dalle Vedove A., Zonta F., Zanforlin E., Demitri N., Ribaudo G., Cazzanelli G., Ongaro A., Sarno S., Zagotto G., Battistutta R., Ruzzene M. and Lolli G. (2020) A novel class of selective CK2 inhibitors targeting its open hinge conformation. Eur. J. Med. Chem. 195, 112267.
Dalle Vedove A., Spiliotopoulos D., D'Agostino V.G., Marchand J.R., Unzue A., Nevado C., Lolli G and Caflisch A. (2018) Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains. ChemMedChem 13, 1479-87.
Marchand J.R., Dalle Vedove A., Lolli G. and Caflisch A. (2017) Discovery of Inhibitors of Four Bromodomains by Fragment-Anchored Ligand Docking. J. Chem. Inf. Model. 57, 2584-97.
Spiliotopoulos D., Wamhoff E.C., Lolli G., Rademacher C. and Caflisch A. (2017) Discovery of BAZ2A bromodomain ligands. Eur. J. Med. Chem. 139, 564-72.
Lolli G., Naressi D., Sarno S. and Battistutta R. (2017) Characterization of the oligomeric states of the CK2 α2β2 holoenzyme in solution. Biochem. J. 474, 2405-16.
Marchand J.R., Lolli G. and Caflisch A. (2016) Derivatives of 3-amino-2-methylpyridine as BAZ2B Bromodomain Ligands: in silico Discovery and in crystallo Validation. J. Med. Chem. 59, 9919-27.
Unzue A., Zhao H., Lolli G., Dong J., Zhu J., Zechner M., Dolbois A., Caflisch A. and Nevado C. (2016) The “gatekeeper” residue modulates the binding mode of acetyl indoles to bromodomains. J. Med. Chem. 59, 3087-97.
Lolli G. and Caflisch A. (2016) High-throughput fragment docking into the BAZ2B bromodomain: Efficient in silico screening for X-ray crystallography. ACS Chem. Biol. 11, 800-7.
