Overview
RNA impacts nearly every aspect of gene expression and it is now clear that the majority of human genetic diseases are caused by mistakes in RNA metabolism. It has become progressively evident that RNA is not just a carrier of genetic information, but also a catalyst and a guide for sequence-specific recognition and processing of other RNA molecules. Progress in our understanding of RNA biology has made it possible to identify RNA molecules as targets of therapeutic intervention and to use RNA as a tool for functional studies and as a novel therapeutic molecule to treat human disease.Since a few years, RNA-based techniques (RNA interference, antisense RNA etc.) are becoming more and more useful in therapy and applied research.
Professor Denti described  her research on RNA Therapies for neurodegenerative disease in an interview by Osservatorio Terapie Avanzate (in italian).
her research on RNA Therapies for neurodegenerative disease in an interview by Osservatorio Terapie Avanzate (in italian).
A recent interview with Prof. Michela A. Denti on advantages and pitfalls of RNA-based therapeutics can be found here(in italian).
Professor Denti spoke about RNA Therapies in a video interviewon RAI 3 (in italian).
Details of the recent participation of the laboratory in the miRNADisEasy consortium, studying microRNAs as lung cancer biomarkers, can be found here(in six languages)
A description of the laboratory ongoing research, its significance and potential applications can be found here.
Simone Detassis talks about "Pitfalls and problems of miRNA expression analysis" in a webinar for the Oligonucleotide Therapeutics Society on February 12, 2019
The laboratory participates in COST Action 17103 "Delivery of Antisense RNA Therapeutics" https://antisenserna.eu
MiRNAQCD: MiRNA Quality Control and Diagnosis
A downloadable R-package to carry out quality control, training and classification analyses on datasets containing MiRNA multiplets.
Research directions
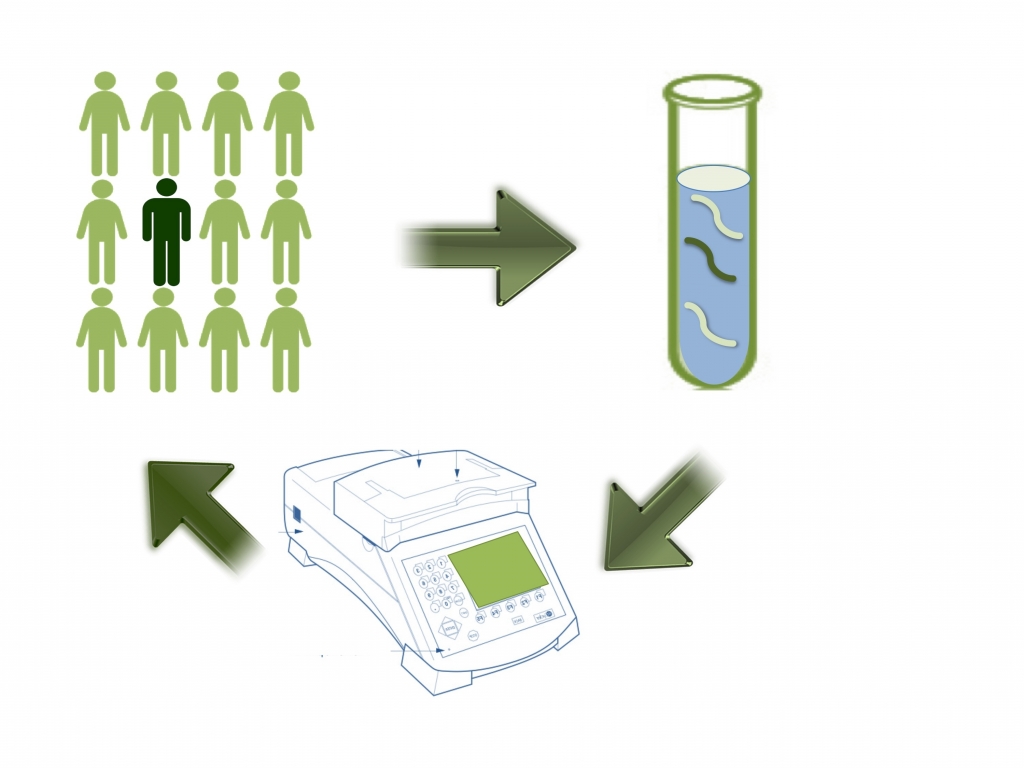
- microRNAs (miRNAs). These are ca. 21 nt regulatory RNAs that control development and differentiation acting as post-transcriptional negative regulators of the expression of key target genes. Recent studies have demonstrated that there is altered expression of miRNA genes in several human malignancies and that miRNAs may act as oncogenes or tumour suppressors. MiRNAs are now also exploited as diagnostic and prognostic biomarkers. We carry out a functional analysis of the miRNAs specifically over- or under-expressed in lung cancers, melanoma, epithelial ovarian cancers and neurodegenerative diseases (e.g. Frontotemporal dementia) search for their target genes and investigate their role in pathogenesis;
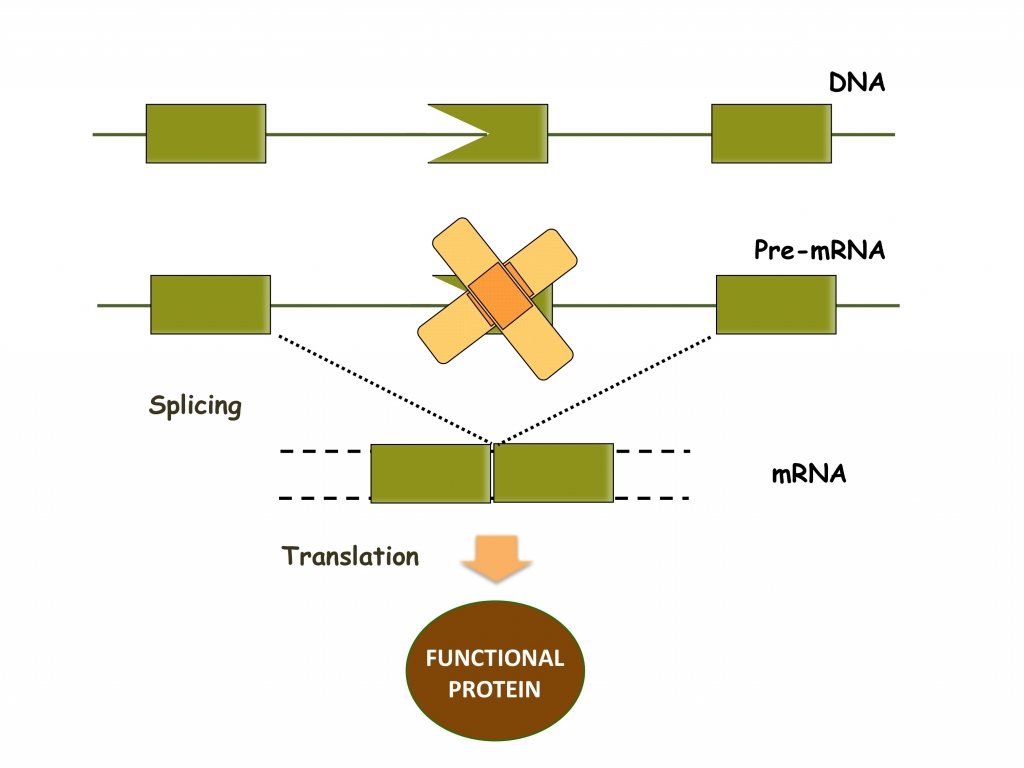
- Antisense RNA-induced exon skipping in gene therapy. The importance of alternative splicing for the diversity of the proteome and the large number of genetic diseases that are due to splicing defects call for methods to modulate alternative splicing decisions. Although splicing can be modulated by antisense oligonucleotides, this approach is confronted with problems of efficient delivery and the need for repeated administrations of large amounts of oligonucleotides. Therefore, we developed a method to induce the efficient and specific skipping of an exon with the help of the U1 small nuclear RNA involved in pre-messenger RNA splicing in the nucleus. Antisense U1 snRNA-induced exon skipping has been successfully employed to devise a therapeutic strategy for Duchenne Muscular Dystrophy. We are now exploring the possibility to apply antisense U1 snRNA-induced exon skipping to other genetic disorders, such as Frontotemporal dementia, retinal dystrophies and metabolic diseases.
Group members
- Michela A. Denti, PI
- Ilaria Brentari, PhD candidate
- Rafael Filipe Battisti Cavichion, PhD candidate
- Elisabetta Callegaro, MSc student
Funding
- European Commission, Marie Skłodowska-Curie Actions - Research and Innovation Staff Exchange – diaRNAgnosis: A novel platform for the direct profiling of circulating cell-free ribonucleic acids in biofluids.
- Bando: PRIN 2022 (D.D. 104/22)
LuCamiR: Development of a microRNA-based test for the screening of lung cancer
Michela Alessandra Denti, Coordinatrice
Codice Protocollo: 20225AZ22F CUP: E53D23012160006
- Bando: PRIN 2022 PNRR (D.D. 1409/22)
MONAD: Combining tailored in silico, in vitro and in vivo models to unveil new NKTR functions associated with rare neurodevelopmental disorders
Michela Alessandra Denti, Responsabile di Unità di Ricerca
Codice Protocollo: P20229PKZC CUP: E53D23015190001

Selected publications
Articles
Covello G, Siva K, Adami V, Denti MA 2023 HCS-Splice: a High-Content Screening Method to Advance the Discovery of RNA Splicing-Modulating Therapeutics. Cells. 2023 Jul 28;12(15):1959. doi: 10.3390/cells12151959.
Piscopo P, Grasso M, Manzini V, Zeni A, Castelluzzo M, Fontana F, Talarico G, Castellano AE, Rivabene R, Crestini A, Bruno G, Ricci L, Denti MA. 2023 Identification of miRNAs regulating MAPT expression and their analysis in plasma of patients with dementia. Frontiers in Molecular Neuroscience 16 doi: 10.3389/fnmol.2023.1127163
Alcantara Santos SA, Frediani Portela LM, Lima Camargo AC, Bessi Constantino F, Thassiani Colombelli K, Naia Fioretto M, Mattos R, De Almeida Fantinatti BE, Denti MA, Piazza S, Felisbino SL, Zambrano E, Justulin LA 2022 miR-18a-5p Is Involved in the Developmental Origin of Prostate Cancer in Maternally Malnourished Offspring Rats: A DOHaD Approach. International Journal of Molecular Sciences. 23:14855. doi: 10.3390/ijms232314855.
Covello G, Ibrahim GH, Bacchi N, Casarosa S*, Denti MA* (2022) Exon Skipping Through Chimeric Antisense U1 snRNAs to Correct Retinitis Pigmentosa GTPase-Regulator (RPGR) Splice Defect. Nucleic Acid Therapeutics doi: 10.1089/nat.2021.0053
Mancini M, Grasso M, Muccillo L, Babbio F, Precazzini F, Castiglioni I, Zanetti V, Rizzo F, Pistore C, De Marino MG, Zocchi M, Del Vescovo V, Licursi V, Giurato G, Weisz A, Chiarugi P, Sabatino L, Denti MA*, Bonapace IM* 2021 DNMT3A epigenetically regulates key microRNAs involved in epithelial-to-mesenchymal transition in prostate cancer. Carcinogenesis. 14:1449-60 doi: 10.1093/carcin/bgab101 *corresponding authors
Simone R, Javad F, Emmett W, Wilkins OG, Almeida FL, Barahona-Torres N, Zareba-Paslawska J, Ehteramyan M, Zuccotti P, Modelska A, Siva K, Virdi GS, Mitchell JS, Harley J, Kay VA, Hondhamuni G, Trabzuni D, Ryten M, Wray S, Preza E, Kia DA, Pittman A, Ferrari R, Manzoni C, Lees A, Hardy JA, Denti MA, Quattrone A, Patani R, Svenningsson P, Warner TT, Plagnol V, Ule J, de Silva R. 2021 MIR-NATs repress MAPT translation and aid proteostasis in neurodegeneration. Nature 594:117–23 doi: 10.1038/s41586-021-03556-6
Fantinatti BEA, Perez ES, Zanella BTT, Valente JS, de Paula TG, Mareco EA, Carvalho RF, Piazza S, Denti MA, Dal-Pai-Silva M 2021 Integrative microRNAome analysis of skeletal muscle of Colossoma macropomum (tambaqui), Piaractus mesopotamicus (pacu), and the hybrid tambacu, based on next-generation sequencing data. BMC Genomics 22, 237. doi: 10.1186/s12864-021-07513-5. https://doi.org/10.1186/s12864-021-07513-5
Castelluzzo M, Perinelli A, Detassis S, Denti MA, Ricci L 2020 MiRNA-QC-and-Diagnosis: An R package for diagnosis based on MiRNA expression.
SoftwareX https://doi.org/10.1016/j.softx.2020.100569.
Detassis S, Del Vescovo V, Grasso M, Masella S, Cantaloni C, Cima L, Cavazza A, Graziano P, Rossi G, Barbareschi M, Ricci L, Denti MA. 2020 miR375-3p distinguishes low-grade neuroendocrine from non-neuroendocrine lung tumors in FFPE samples. Front. Mol. Biosci. 7:86. doi: 10.3389/fmolb.2020.00086
Masè M, Grasso M, Avogaro L, Nicolussi Giacomaz N, D’Amato E, Tessarolo F, Graffigna A, Denti MA*, Ravelli F*. 2019 Upregulation of miR-133b and miR-328 in Patients With Atrial Dilatation: Implications for Stretch-Induced Atrial Fibrillation. Front Physiol 10:1133 doi: 10.3389/fphys.2019.01133 *equally contributing
Detassis S, Grasso M, Tabraue-Chávez M, Marin Romero A, Ilyine H, Ress C, Ceriani S, Erspan M, Maglione A, Díaz-Mochón J, Pernagallo S*, Denti MA* 2019 A new platform for the direct profiling of microRNAs in biofluids. Analytical Chemistry. 91:5874-5880 doi: 10.1021/acs.analchem.9b00213 *equally contributing
Grasso M, Piscopo P, Talarico G, Ricci L, Crestini A, Tosto G, Gasparini M, Bruno G, Denti MA*, Confaloni A* 2019 Plasma microRNAs profiling distinguishes patients with frontotemporal dementia from healthy subjects. Neurobiology of Aging 84:240.e1-e12 doi: 10.1016/j.neurobiolaging.2019.01.024 *equally contributing
Piscopo P, Grasso M, Puopolo M, D’Acunto E, Talarico G, Crestini A, Gasparini M, Campopiano R, Gambardella S, Castellano AE, Bruno G, Denti MA*, Confaloni A* 2018 Circulating miR-127-3p as a biomarker for differential diagnosis in Frontotemporal Dementia. J. Alzheimer’s Disease 65: 455-464 doi: 10.3233/JAD-180364 *equally contributing
Pistore C, Giannoni E, Colangelo T, Rizzo F, Magnani E, Muccillo L, Giurato G, Mancini M, Rizzo S, Riccardi M, Sahnane N, Del Vescovo V, Kishore K, Mandruzzato M, Macchi F, Pelizzola M, Denti MA, Furlan D, Weisz A, Colantuoni V, Chiarugi P, Bonapace IM 2017 DNA methylation variations are required for epithelial-to-mesenchimal transition induced by cancer associate fibroblasts in prostate cancer cells Oncogene 36:5551-6 doi: 10.1038/onc.2017.159
Masè M, Grasso M, Avogaro L, D'Amato E, Tessarolo F, Graffigna A, Denti MA*, Ravelli F* 2017 Selection of reference genes is critical for miRNA expression analysis in human cardiac tissue. A focus on atrial fibrillation. Scientific Reports 7:41127 doi: 10.1038/srep41127 *equally contributing
Ferri L, Covello G, Caciotti A, Guerrini R, Denti MA*, Morrone A* 2016 Double-target antisense U1snRNAs correct mis-splicing due to GLA deep intronic mutations in Fabry Disease Molecular Therapy Nucleic Acids 5:e380 doi: 10.1038/mtna.2016.88 *equally contributing
Moncini M, Lunghi M Valmadre A, Grasso M, Del Vescovo V, Riva P, Denti MA, Venturin M 2016 The miR-15/107 family of microRNA genes regulates CDK5R1/p35 with implications for Alzheimer’s disease pathogenesis Mol Neurobiol. doi: 10.1007/s12035-016-0002-4
Piscopo P, Grasso M, Fontana F, Crestini A, Puopolo M, Del Vescovo V, Venerosi A, Calamandrei G, Vencken SF, Greene CM, Confaloni A, Denti MA 2016 Reduced miR-659-3p levels correlate with progranulin increase in hypoxic conditions: implications for frontotemporal dementia Front Mol Neurosci 9:31 doi: 10.3389/fnmol.2016.00031
Ricci L, Del Vescovo V, Cantaloni C, Grasso M, Barbareschi M, Denti MA 2015 Statistical analysis of a Bayesian classifier based on the expression of miRNAs BMC Bioinformatics 16:287 doi: 10.1186/s12859-015-0715-9
Bacchi N, Messina A, Burtscher V, Dassi E, Provenzano G, Bozzi Y, Demontis GC, Koschak A*, Denti MA*, Casarosa S* 2015 A new splicing isoform of Cacna2d4 mimicking the effects of c.2451insC mutation in the retina. Novel molecular and electrophysiological insights. Invest Ophthalmol Vis Sci. 56:4846-56. doi: 10.1167/iovs.15-16410 *equally contributing
Covello G, Siva K, Adami V, Denti MA 2014 An electroporation protocol for efficient DNA transfection in PC12 cells.” Cytotechnology 66:543-553 doi: 10.1007/s10616-013-9608-9
Bisio A, De Sanctis V, Del Vescovo V, Denti MA, Jegga AG, Inga A, Ciribilli Y 2013 Identification of new p53 target microRNAs by bioinformatics and functional analysis” BMC Cancer 13:552 doi: 10.1186/1471-2407-13-552
Del Vescovo V, Meier T, Inga A, Denti MA*, Borlak J* 2013 A Cross-Platform Comparison of Affymetrix and Agilent Microarrays Reveals Discordant miRNA Expression in Lung Tumors of c-Raf Transgenic Mice PLoS ONE 8: e78870. doi: 10.1371/journal.pone.0078870 *equally contributing
Barbareschi M, Cantaloni C, Del Vescovo V, Cavazza A, Monica V, Carella R, Rossi G, Morelli L, Cucino A, Silvestri M, Tirone G, Pelosi G, Graziano P, Papotti M, Dalla Palma P, Doglioni C, Denti MA 2011 Heterogeneity of large cell carcinoma of the lung: an immunophenotypic and miRNA based analysis Am. J. Clin. Pathol. 136:773-782 doi: 10.1309/AJCPYY79XAGRAYCJ
Del Vescovo V, Cantaloni C, Cucino A, Girlando S, Silvestri M, Bragantini E, Fasanella S, Cuorvo LV, Dalla Palma P, Rossi G, Papotti M, Pelosi G, Graziano P, Cavazza A, Denti MA, Barbareschi M 2011 miR-205 expression levels in non-small cell lung cancer do not always distinguish adenocarcinomas from squamous cell carcinomas Am. J. Surg. Pathol. 35:268-275 doi: 10.1097/PAS.0b013e3182068171
Reviewes
Brentari I, Zadorozhna M, Denti MA, Giorgio E 2023 RNA therapeutics for neurological diseases, British Medical Bulletin. ldad010, doi: 10.1093/bmb/ldad010
Brancato V, Brentari I, Coscujuela Tarrero L, Furlan M, Nicassio F*, Denti MA* 2022 News from around the RNA world: new avenues in RNA biology, biotechnology and therapeutics from the 2022 SIBBM meeting. Biology open 11(10) doi:10.1242/bio.059597 *equally contributing
Hammond SM, Aartsma‐Rus A, Alves S, Borgos SE, Buijsen RAM, Collin RWJ, Covello G, Denti MA, Desviat LR, Echevarría L, Foged C, Gaina G, Garanto A, Goyenvalle AT, Guzowska M, Holodnuka I, Jones DR, Krause S, Lehto T, Montolio M, Van Roon‐Mom W, Arechavala‐Gomeza V 2021 Delivery of oligonucleotide‐based therapeutics: challenges and opportunities. EMBO Mol Med 13: e13243. doi:10.15252/emmm.202013243
Precazzini F, Detassis S, Imperatori AS, Denti MA*, Campomenosi P* 2021 Measurements methods for the development of microRNA-based tests for cancer diagnosis Int. J. Mol. Sci. 22:1176 doi:10.3390/ijms22031176 *equally contributing
Detassis S, Grasso M, Del Vescovo V, Denti MA 2017 microRNAs Make the Call in Cancer Personalized Medicine Front. Cell Dev. Biol. 5:86 doi:10.3389/fcell.2017.00086
Godfrey C, Desviat LR, Smedsrod B, Piétri-Rouxel F, Denti MA, Disterer P, Lorain S, Nogales-Gadea G, Sardone V, Anwar R, El Andaloussi S, Lehto T, Khoo B, Brolin Hjortkjaer C, van Roon-Mom WMC, Goyenvalle A, Aartsma-Rus A, Arechavala-Gomeza V 2017 Delivery is key: lessons learnt from developing splice switching antisense therapy EMBO Molecular Medicine doi: 10.15252/emmm.201607199
Fontana F, Siva K, Denti MA 2015 A network in Fronto Temporal Dementia - an RNA perspective Front Mol Neurosci 8:9 doi: 10.3389/fnmol.2015.00009
Grasso M, Piscopo P, Confaloni A, Denti MA 2014 Circulating miRNAs as biomakers for neurodegenerative diseases Molecules 19:6891-6910 doi:10.3390/molecules19056891
Bacchi N, Casarosa S*, Denti MA* 2014 Splicing-correcting therapeutic approaches for retinal dystrophies: where endogenous gene regulation and specificity matter Invest Ophthalmol Visual Sci. 55:3285-94 doi: 10.1167/iovs.14-14544 *equally contributing
Del Vescovo V, Grasso M, Barbareschi M, Denti MA 2014 microRNAs as lung cancer biomarkers World Journal Clinical Oncology 5:604-620 doi: 10.5306/wjco.v5.i4.604
Siva K, Covello G, Denti MA 2014 Exon-skipping Antisense Oligonucleotides to Correct Missplicing in Neurogenetic Diseases” Nucl. Acids Therap. 24: 69-86 doi: 10.1089/nat.2013.0461
Book chapters
Zeni A, Grasso M, Denti MA 2021 Identification of miRNAs Bound to an RNA of Interest by MicroRNA Capture Affinity Technology (miR-CATCH). in “Post-Transcriptional Gene Regulation” Methods in Molecular Biology. Erik Dassi (Ed.) Springer ISBN 978-1-0716-1850-9
Denti MA and Covello G 2017 “Oligonucleotide therapy” in “Safety and Efficacy of Gene-Based Therapeutics for Inherited Disorders” Nicola Brunetti-Pierri (Ed.) Springer ISBN 978-3-319-53455-8 doi: 10.1007/978-3-319-53457-2_9
Del Vescovo V, Denti MA 2015 “microRNA and lung cancer” in “microRNA: Cancer. From molecular biology to clinical practice” Gaetano Santulli (Ed.) Springer ISBN 978-3-319-23729-9
Grasso M, Piscopo P, Crestini A, Confaloni A, Denti MA 2015 “Circulating microRNAs in neurodegenerative diseases” in “Circulating microRNAs in disease diagnostics and their potential biological relevance” Peter Igaz (Ed.) Springer ISBN 978-3-0348-0953-5
Grasso M, Fontana F, Denti MA 2015 “Circulating cell-free microRNAs as biomarkers for neurodegenerative diseases” in “Frontiers in Neurotherapeutics: Mapping Nervous System Diseases via microRNAs” Christian Barbato and Francesca Ruberti (Eds.) CRC Press-Taylor & Francis Group (Boca Raton FL, USA) ISBN 978148226352

