Overview
Our research focuses on understanding the neurobiological mechanisms underlying neuropsychiatric disorders including autism, fragile X syndrome, and schizophrenia.
Recent findings indicate that these neuropsychiatric disorders might arise from the alteration of normal brain developmental trajectories. Although most research on sensitive periods has been focused on early childhood, examining the adolescent brain changes might represent a major opportunity for autism treatment. Adolescence is a time of tremendous brain reorganization and plasticity, particularly in brain regions involved in higher-level cognitive processes such as learning, social and emotional behaviour.
Our laboratory aims to understand how adolescence impacts brain and circuit functions in experimental models of neuropsychiatric disorders. We employ a multidisciplinary approach combining functional genomic approaches, molecular and behavioural studies to develop a more comprehensive understanding of the relationship between gene dysfunctions, brain circuits, and behaviour.
Research directions
To dissect convergent and distinct transcriptomic dysfunctions across different genetic aetiologies
Brain transcriptome studies are emerging as a strategic tool to uncover functionally relevant alterations in neuropsychiatric disorders. We are currently investigating whether common transcriptional abnormalities during sensitive periods of postnatal brain development have long-term consequences on brain functions. Our ultimate goal is the identification of druggable targets, from the integrative analysis of transcriptomic data. This could potentially lead to the development of novel treatments, with the advantage of creating the premises for therapeutic targets with a window for early treatment.
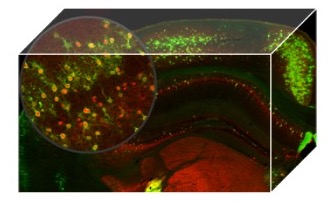 To map whole-brain activation evoked by social behaviour at cellular resolution
To map whole-brain activation evoked by social behaviour at cellular resolution
A major challenge in neuroscience is to understand how brain activation produces and mediates social behaviours. We approach this question by combining imaging technologies with genetic strategies such as “FosTRAP system” to label activated neurons following a social experience. The main goal is to investigate the molecular mechanisms by which these activated neuronal cells exert their behavioural effects.
Group members
We are looking for motivated candidates, including undergraduate, master, and doctoral students to join our lab. If you are interested in Molecular and Behavioural Neuroscience, please contact giovanni.provenzano [at] unitn.it
Collaborations
- Flavia Antonucci, University of Milan, Milano, Italy
- Silvano Piazza, ICGEB, Trieste, Italy
- Susanna Pietropaolo, University of Bordeaux, Bordeaux Cedex, France
- Sara Bonini, University of Brescia, Brescia, Italy
- Josep Rubert, Wageningen University, Wageningen, The Netherlands
- Yuri Bozzi, CIMeC - Center for Mind/Brain Sciences, University of Trento, Italy
Molecular and Behavioural Neuroscience laboratory is part of TRAIN – “Trentino Autism Initiative” https://projects.unitn.it/train/
Selected publications
Pietropaolo S, Provenzano G. Targeting Excitation-Inhibition Imbalance in Neurodevelopmental and Autism Spectrum Disorders. Front Neurosci. 2022 Jul 8;16:968115. doi: 10.3389/fnins.2022.968115. eCollection 2022.
Focchi E, Cambria C, Pizzamiglio L, Murru L, Pelucchi S, D'Andrea L, Piazza S, Mattioni L, Passafaro M, Marcello E, Provenzano G, Antonucci F (2022) ATM rules neurodevelopment and glutamatergic transmission in the hippocampus but not in the cortex. Cell Death Dis, 13(7):616, doi:10.1038/s41419-022-05038-7.
Balasco L, Pagani M, Pangrazzi L, Chelini G, Viscido F, Chama AGC, Galbusera A, Provenzano G, Gozzi A, Bozzi Y (2022) Somatosensory cortex hyperconnectivity and impaired whisker-dependent responses in Cntnap2. Neurobiol Dis, 169:105742, doi:10.1016/j.nbd.2022.105742.
Zerbi V, Pagani M, Markicevic M, Matteoli M, Pozzi D, Fagiolini M, Bozzi Y, Galbusera A, Scattoni ML, Provenzano G, Banerjee A, Helmchen F, Basson MA, Ellegood J, Lerch JP, Rudin M, Gozzi A, Wenderoth N. Brain mapping across 16 autism mouse models reveals a spectrum of functional connectivity subtypes. Mol Psychiatry. 2021 Aug 11. doi: 10.1038/s41380-021-01245-4.
Balasco L, Chelini G, Bozzi Y, Provenzano G. Whisker Nuisance Test: A Valuable Tool to Assess Tactile Hypersensitivity in Mice. Bio Protoc. 2019 August 20, Vol 9, Iss 16. doi: 10.21769/BioProtoc.3331.
Chelini G, Zerbi V, Cimino L, Grigoli A, Markicevic M, Libera F, Robbiati S, Gadler M, Bronzoni S, Miorelli S, Galbusera A, Gozzi A, Casarosa S, Provenzano G*, Bozzi Y*. Aberrant Somatosensory Processing and Connectivity in Mice Lacking Engrailed-2. J Neurosci. 2019 Feb 20;39(8):1525-1538. doi: 10.1523/JNEUROSCI.0612-18.2018.
Provenzano G#, Corradi Z, Monsorno K, Fedrizzi T, Ricceri L, Scattoni ML, Bozzi Y. Comparative Gene Expression Analysis of Two Mouse Models of Autism: Transcriptome Profiling of the BTBR and En2 (-/-) Hippocampus. Front Neurosci. 2016 Aug 25;10:396. doi: 10.3389/fnins.2016.00396.
Provenzano G, Pangrazzi L, Poli A, Sgadò P, Genovesi S, Zunino G, Berardi N, Casarosa S, Bozzi Y (2014). Hippocampal dysregulation of neurofibromin-dependent pathways is associated with impaired spatial learning in Engrailed 2 knockout mice. J Neurosci. 2014 Oct 1;34(40):13281-8. doi: 10.1523/JNEUROSCI.2894-13.2014.
Chabrol E, Navarro V, Provenzano G, Cohen I, Dinocourt C, Rivaud-Péchoux S, Fricker D, Baulac M, Miles R, LeGuern E and Baulac S (2010). Electro-clinical Characterization of Epileptic Seizures in LGI1-deficient Mice. Brain. 2010 Sep;133(9):2749-62. doi: 10.1093/brain/awq171. Epub 2010 Jul 21.
* = equal contribution
