Overview
Epigenetics allows cells to adapt to environmental alterations, causing heritable yet reversible changes of the cellular state. Perturbations of the epigenetic machinery are often responsible for diseases. The focus of our team is to investigate how epigenetic changes could impact on stem cells, both in physiological and pathological settings. We aim to dissect the molecular mechanisms through which chromatin players mediate the epigenetic responses to environmental signals in stem cells. We intent to tackle this biological question using a multidisciplinary approach that by combining dynamic single cell tracking strategy with the epigenetic profiling of cells, should allow to determine the network of combinatorial epigenetic modifications that establish a stem cell-like epigenetic state. On the same line, we are also interested in understanding the contribution of oncogene-induced epigenetic reprogramming to tumor progression and metastasis formation.
Research directions
-
Deciphering the role of epigenetic reprogramming in driving cell plasticity during tumor progression and metastasis
Cancer genomics showed that tumor progression to metastasis formation is poorly supported by further genetic alterations, implying that the adaptation capacity of disseminating tumor cells to foreign microenvironments could rely on reversible epigenetic alterations. Using a pre-clinical model of triple negative breast cancer, we are investigating the contribution of epigenetic reprogramming to cancer-driven cell plasticity. We are asking how the oncogenic epigenetic state fosters cancer cell fitness during tumor progression by preserving the memory of previously encountered stimulus through self-propagating molecular signatures. To address these biological questions, we integrate genome-wide profiling of chromatin state with the 3D genome topology. We are developing computational and experimental approaches to untangle the contribution of the multiple layers of epigenetic information in ensuring epigenetic memory in cancer cells. As an orthogonal approach, we are implementing the live imaging methodology to dissect the role of mitotic bookmarking in preserving the epigenetic state at enhancers and other cis-regulatory elements.
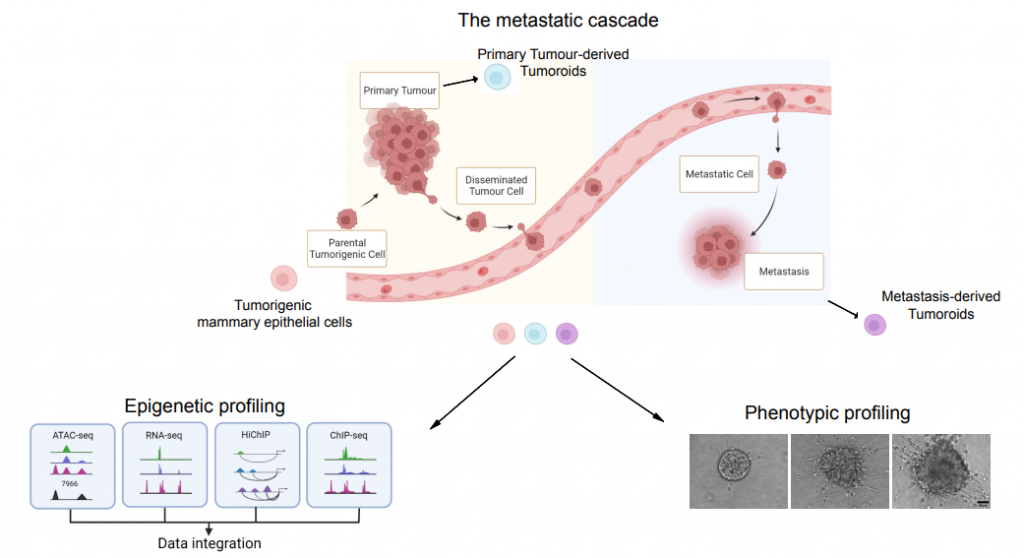
-
Dissecting the interplay between chromatin condensates and mechanobiology in rare genetic disorders
Chromatin is organized in biomolecular condensates that compartmentalize the genome function and control gene expression. We aim to solve chromatin domains by super-resolution imaging and optogenetic-based tools, to determine the contribution of chromatin compartmentalization and nuclear architecture to the onset and progression of rare genetic disease. Many inherited disorders result from mutations in the genes encoding various components of the epigenetic machinery. Among others, we are investigating the pathological role of the mutated MLL4 in the etiology of the Kabuki Syndrome (KS), a multiple congenital anomaly disorder lacking any therapeutic options. By developing an in vitro disease model system, we are establishing whether the pathogenesis of this disease depends on the MLL4-associatd epigenetic alterations. By using cutting-edge technologies, we are developing new approaches to define the role of chromatin condensates in establishing the nuclear mechanical properties to then exploit novel therapeutic approach to treat KS. In this respect, we are interested in dissecting the mechanisms by which the intrinsically disorder regions (IDRs) of MLL4 guide the assembly of transcriptional condensates.
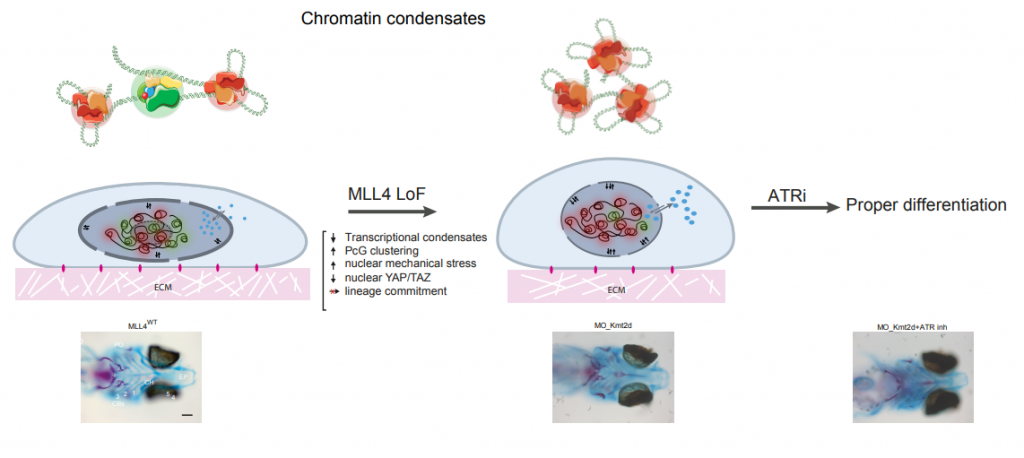
Group members
- Alessio Zippo, PI
- Sven Beyes, Post-doc
- Silvia Scalisi, Post-doc
- Sara Lago, Post-doc
- Daniela Michelatti, Lab manager
- Sarah D’Annunzio, PhD student
- Chiara Bernardis, PhD student
- Maria Luce Negri, PhD student
- Leonardo Norelli, PhD student
- Lisa Fol, undergraduate student
- Nicole Cona, undergraduate student
- Antonio Timpone, undergraduate student
We are currently looking for highly motivated candidates for a PhD and Post-doc position interested in investigating the role of chromatin compartmentalization in diseases.
Please directly contact the PI, Alessio Zippo
Collaborations
- Prof. Alvaro Rada Iglesias (IBBTEC, Spain)
- Prof. Giorgio Stassi (Università degli Studi di Palermo, Italy)
- Prof. Matilde Todaro (Università degli Studi di Palermo, Italy)
- Mark Neil (Imperial College, UK)
- Prof. Vania Broccoli (San Raffaele Scientific Institute, Italy)
- Prof. Andrea Bassi (Politecnico di Milano, Italy)
- Prof. Ruocco Giancarlo (University of Rome, Italy)
- Prof. Giangaetano Tartaglia (University of Rome, Italy)
- Dr. Paolo Maiuri (IFOM Institute, Italy)
Funding
Bando: PRIN 2022 (D.D. 104/22)
Dissecting the interplay between chromatin-associated condensates and nuclear mechanical properties in Kabuki Syndrome
Alessio Zippo, Coordinatore
Codice Protocollo: 2022H79275 CUP: E53D23009880001

Selected publications
MLL4-associated condensates counterbalance Polycomb-mediated nuclear mechanical stress in Kabuki syndrome. Fasciani A, D'Annunzio S, Poli V, Fagnocchi L, Beyes S, Michelatti D, Corazza F, Antonelli L, Gregoretti F, Oliva G, Belli R, Peroni D, Domenici E, Zambrano S, Intartaglia D, Settembre C, Conte I, Testi C, Vergyris P, Ruocco G, Zippo A.
Nat Genet. 2020 Dec;52(12):1397-1411
GADD34 is a modulator of autophagy during starvation. Gambardella G, Staiano L, Moretti MN, De Cegli R, Fagnocchi L, Di Tullio G, Polletti S, Braccia C, Armirotti A, Zippo A, Ballabio A, De Matteis MA, di Bernardo D.
Sci Adv. 2020 Sep 25;6(39):eabb0205.
High-throughput 3D imaging of single cells with light-sheet fluorescence microscopy on chip. Sala F, Castriotta M, Paiè P, Farina A, D'Annunzio S, Zippo A, Osellame R, Bragheri F, Bassi A.
Biomed Opt Express. 2020 Jul 17;11(8):4397-4407.
Truncated BRPF1 cooperates with Smoothened to promote adult Shh medulloblastoma. Aiello G, Ballabio C, Ruggeri R, Fagnocchi L, Morassut I, Caron D, Francesca Garilli, Gianno F, Giangaspero F, Piazza S, Romanel A, Zippo A and Tiberi L.
Cell Rep. 2019 Dec 17;29(12):4036-4052.e10
SETD5 regulates chromatin methylation state and preserves global transcriptional fidelity during brain development and neuronal wiring Sessa A, Fagnocchi L, Mastrototaro G, Massimino L, Zaghi M, Indrigo M, Cattaneo S, Martini D, Gabellini C, Pucci C, Fasciani A, Belli R, Taverna S, Andreazzoli M, Zippo A*, Broccoli V*.
Neuron 2019 Oct 23;104(2):271-289.e13 * Co-senior Authors.
The proneural gene ASCL1 governs the transcriptional subgroup affiliation in glioblastoma stem cells by directly repressing the mesenchymal gene NDRG1. Narayanan A, Gagliardi F, Gallotti AL, Mazzoleni S, Cominelli M, Fagnocchi L, Pala M, Piras IS, Zordan P, Moretta N, Tratta E, Brugnara G, Altabella L, Bozzuto G, Gorombei P, Molinari A, Padua RA, Bulfone A, Politi LS, Falini A, Castellano A, Mortini P, Zippo A, Poliani PL, Galli R.
Cell Death Differ. 2018 Dec 11.
Tumorigenic Cell Reprogramming and Cancer Plasticity: Interplay between Signaling, Microenvironment, and Epigenetics. Vittoria Poli, Luca Fagnocchi and Alessio Zippo.
Stem Cell Int. 2018 May 2;2018:4598195.
Enhancer reprogramming in tumor progression: a new route towards cancer cell plasticity. Fagnocchi L, Poli V, Zippo A.
Cell Mol Life Sci. 2018 Apr 24
MYC-driven epigenetic reprogramming favors the onset of tumorigenesis by inducing a stem cell-like state. Poli V, Fagnocchi L, Fasciani A, Cherubini A, Mazzoleni S, Ferrillo S, Miluzio A, Gaudioso G, Vaira V, Turdo A, Giaggianesi M, Chinnici A, Lipari E, Bicciato S, Bosari S, Todaro M, Zippo A.
Nat Commun. 2018 Mar 9;9(1):1024
A cytosolic Ezh1 isoform modulates a PRC2-Ezh1 epigenetic adaptive response in potmitotic cells. Bodega B, Marasca F, Ranzani V, Cherubini A, Della Valle F, Neguembor MV, Wassef M, Zippo A, Lanzuolo C, Pagani M, Orlando V.
Nat Struct Mol Biol. 2017 May; 24(5):444-452
Fluorescence Resonance Energy Transfer microscopy for measuring chromatin complex structure and dynamics. Alessandro Cherubini and Alessio Zippo.
Methods in Molecular Biology, 2016
Integration of Signalling pathways with the epigenetic machinery in the maintenance of stem cells. Luca Fagnocchi, Mazzoleni Stefania and Alessio Zippo.
Stem Cell International, March 2016.
Lamin A/C sustains PcG proteins architecture maintaining repression of target genes. Cesarini E, Mozzetta C, Marullo F, Gregoretti F, Gargiulo A, Columbaro M, Cortesi A, Antonelli L, Di Pelino S, Squarzoni S, Palacios D, Zippo A, Bodega B, Oliva G, Lanzuolo C.
Journal of Cell Biology 2015 Nov 9;211(3):533-51
FOSL1 controls the assembly of endothelial cells into capillary tubes by direct repression of v and 3 integrin transcription. Evellin S, Galvagni F, Zippo A, Neri F, Orlandini M, Incarnato D, Dettori D, Neubauer S, Kessler H, Wagner EF, Oliviero S.
Mol Cell Biol. 2013 Mar;33(6):1198-209.
MYC regulates the transcription of PRC2 to control the expression of developmental genes in embryonic stem cells. Francesco Neri, Alessio Zippo, Anna Krepelova, Alessandro Cherubini, Marina Rocchigiani, and Salvatore Oliviero.
Mol Cell Biol. 2012 Feb;32(4):840-5
Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S.
Cell. 2009 Sep 18; 138(6):1122-36.
PIM1-dependent phosphorylation of Histone H3 at Serine 10 is required for the transcriptional activation of MYC-target genes. Zippo A, De Robertis A, Serafini R, Oliviero S.
Nat Cell Biol. 2007 Aug; 9(8):932-44.
