
Overview
Cancer is an extremely widespread, heterogeneous, and still too often lethal disease. Some forms of cancer are associated with the age of the individual, or the gender, others with the exposure to environmental factors, some are extremely aggressive, others indolent. All indiscriminately, however, have a genetic basis and molecular aberrations that distinguish them. Defining the molecular mechanisms underlying neoplastic processes is a crucial goal for improving clinical decisions, treatment efficacy, and ultimately patient survival. Primary interest of my research group is the generation of in vitro and in vivo study models that recapitulate the different stages of tumorigenesis to understand the molecular circuits responsible for the onset, aggressiveness and resistance to treatment of different neoplastic diseases. Such models are an indispensable tool for the identification of novel markers of early stages of disease, effective therapeutic targets, as well as for pre-clinical and co-clinical evaluation of the efficacy of new personalized cancer treatments.
Research directions
-
Extracellular signaling in prostate tissue homeostasis and tumorigenesis
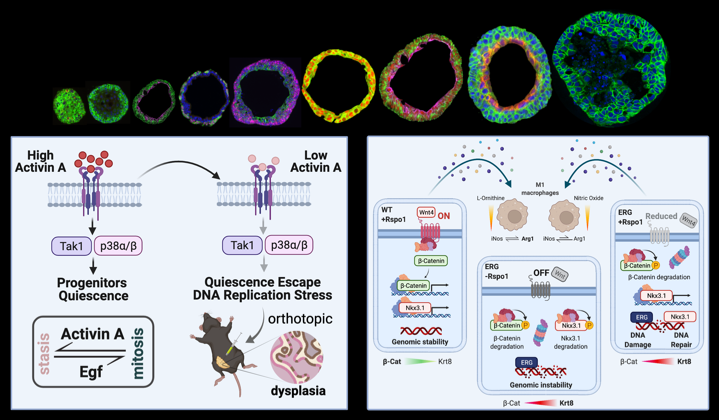
Few secreted factors, mainly proteins, have been characterized in human prostate cancer, but their relevance to tumor onset, progression and response to specific treatments is not yet well understood. Our major interest lies in taking advantage of prostate organoids and cell lines to explore the roles of secreted molecules in prostate tissue homeostasis and cancer pathogenesis.
(Cambuli, Foletto et al., 2022; Lorenzoni, De Felice et al., 2022). -
Identification of novel targeted therapy improving clinical protocols
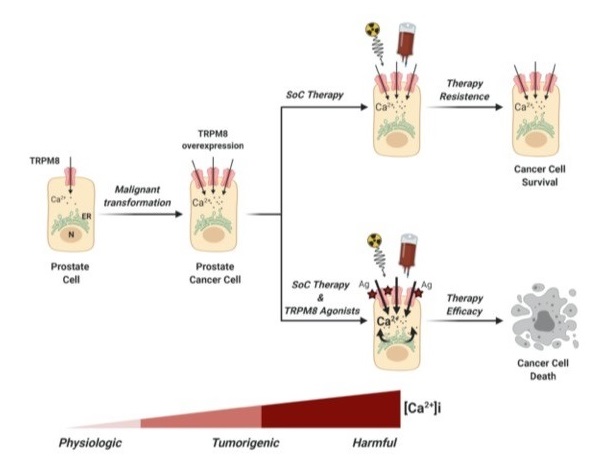
We and other groups have identified Transient Receptor Potential Cation Channel Melastatin Member 8 (TRPM8) as a novel potential candidate for therapeutic approaches of precision oncology to different forms of solid tumors. In vitro experiments on human and mouse cancer cell lines mirroring aggressive forms of cancer, have demonstrated how pharmacological targeting of TRPM8 can improve the efficacy of standard-of-care protocols. Preclinical in vivo study with innovative solution of drug delivery based on chimeric molecules is the next goal of our lab.
(Alaimo et al., 2020; Genovesi et al., 2022). -
Prostate Cancer Inflammation and Immunology
Genome instability in prostate cancer is largely dependent by genomic rearrangements rather than single nucleotide mutations, which are rare events. Because of this, prostate cancer cells are poorly decorated by classic neo-antigens and badly intercepted by the immune system. Besides neo-antigens, a second class of immunogenic elements are the so-called Tumor Associated Antigens. TAA may originate, for example, from altered RNA splicing, post-translational modifications or degradation of wild type proteins, or ectopic expression of embryonal and germline genes. TAA identification is a major interest of our group, the finale goal in this case is the generation and pre-clinical testing of TAA-based vaccines for the treatment of prostate cancer.
Team members
- Andrea Lunardi, PI
- Alessandro Alaimo, Senior PostDoc
- Nicole Anesi PreDoc
- Sacha Genovesi, Research Assistant
Awards
- 2008- Begnudelli Award, Pezcoller-AACR Foundation, Trento, Italy.
- 2018-Begnudelli Award, Pezcoller-AACR Foundation, Trento, Italy.
- 2014- Career Development Award, The Giovanni Armenise Harvard Foundation.
- 2018-EMBO Molecular Medicine Award-EMBO/EMBL Organoid Symposium.
Grants
- Armenise/Harvard Career Development Award (Past)
- MIUR/PRIN 2017 (Active)
- Lega Italiana per la Lotta ai Tumori-LILT (Active)
- Fondazione Trentina per la Ricerca sui Tumori (Active)
- AIRC (Active)
Collaborators
- Nicola Ternette, Nuffield Department of Medicine, University of Oxford, UK.
- Massimo Loda, Weill Cornell Medicine, New York, USA.
- Michael M. Shen, Columbia University New York, USA.
- Marianna Kruithof-de Julio, Bern University, Bern, Switzerland.
- Martin G. Pomper, Johns Hopkins Medicine, Baltimore, USA.
- Alessandro Romanel, University of Trento, Trento, Italy.
- Francesca Demichelis, University of Trento, Trento, Italy.
- Mattia Barbareschi, Santa Chiara Hospital, Trento, Italy.
- Orazio Caffo, Santa Chiara Hospital, Trento, Italy.
- Alberto Briganti, San Raffaele Hospital, Milan, Italy.
- Claudio Doglioni, San Raffaele Hospital, Milan. Italy.
- Rodolfo Montironi, Ospedali Riuniti, Ancona, Italy.
- Maurizio Taglialatela, University of Naples Federico II, Naples, Italy.
- Valeria Poli, University of Turin, Turin, Italy.
- Marco Gaspari, University of Catanzaro, Catanzaro, Italy.
- Licio Collavin, University of Trieste, Trieste, Italy.
- Giannino Del Sal, ICGEB-University of Trieste, Trieste, Italy.
Funding
Bando: PRIN 2022 (D.D. 104/22)
Industrial Biocatalysts Based on Outer Membrane Vesicles
Andrea Lunardi, Responsabile di Unità
Codice Protocollo: 202248L9YA CUP: E53D23010540006

Selected publications
Articles
Lorenzoni M, De Felice D, Beccaceci G, Di Donato G, Foletto V, Genovesi S, Bertossi A, Cambuli F, Lorenzin F, Savino A, Avalle L, Cimadamore A, Montironi R, Weber V, Carbone FG, Barbareschi M, Demichelis F, Romanel A, Poli V, Del Sal G, Julio MK, Gaspari M, Alaimo A, Lunardi A. 2022 ETS-related gene (ERG) undermines genome stability in mouse prostate progenitors via Gsk3β dependent Nkx3.1 degradation. Cancer Lett. 2022 May 28;534:215612. doi: 10.1016/j.canlet.2022.215612. Epub 2022 Mar 5. PMID: 35259458
Cambuli F, Foletto V, Alaimo A, De Felice D, Gandolfi F, Palumbieri MD, Zaffagni M, Genovesi S, Lorenzoni M, Celotti M, Bertossio E, Mazzero G, Bertossi A, Bisio A, Berardinelli F, Antoccia A, Gaspari M, Barbareschi M, Fiorentino M, Shen MM, Loda M, Romanel A, Lunardi A. 2022 Intra-epithelial non-canonical Activin A signaling safeguards prostate progenitor quiescence. EMBO Rep. 2022 May 4;23(5):e54049. doi: 10.15252/embr.202154049. Epub 2022 Mar 7. PMID: 35253958
Genovesi S, Moro R, Vignoli B, De Felice D, Canossa M, Montironi R, Carbone FG, Barbareschi M, Lunardi A, Alaimo A. 2022 Trpm8 Expression in Human and Mouse Castration Resistant Prostate Adenocarcinoma Paves the Way for the Preclinical Development of TRPM8-Based Targeted Therapies. Biomolecules. 2022 Jan 23;12(2):193. doi: 10.3390/biom12020193.PMID: 35204694
Alaimo A, Lorenzoni M, Ambrosino P, Bertossi A, Bisio A, Macchia A, Zoni E, Genovesi S, Cambuli F, Foletto V, De Felice D, Soldovieri MV, Mosca I, Gandolfi F, Brunelli M, Petris G, Cereseto A, Villarroel A, Thalmann G, Carbone FG, Kruithof-de Julio M, Barbareschi M, Romanel A, Taglialatela M, Lunardi A. 2020 Calcium cytotoxicity sensitizes prostate cancer cells to standard-of-care treatments for locally advanced tumors. Cell Death Dis. Dec 7;11(12):1039. doi: 10.1038/s41419-020-03256-5. PMID: 33288740
Bezzi M, Seitzer N, Ishikawa T, Reschke M, Chen M, Wang G, Mitchell C, Ng C, Katon J, Lunardi A, Signoretti S, Clohessy JG, Zhang J, Pandolfi PP. 2018 Diverse genetic-driven immune landscapes dictate tumor progression through distinct mechanisms. Nat Med. Feb;24(2):165-175. doi: 10.1038/nm.4463. Epub 2018 Jan 8. PMID: 29309058
Lunardi A, Varmeh S, Chen M, Taulli R, Guarnerio J, Ala U, Seitzer N, Ishikawa T, Carver BS, Hobbs RM, Quarantotti V, Ng C, Berger AH, Nardella C, Poliseno L, Montironi R, Castillo-Martin M, Cordon-Cardo C, Signoretti S, Pandolfi PP. 2015. Suppression of CHK1 by ETS family members promotes DNA damage response by-pass and tumorigenesis. Cancer Discov. Feb 4. pii: CD-13-1050. PMID:25653093
Guarnerio J, Riccardi L, Taulli R, Maeda T, Wang W, Hobbs RM, Song MS, Sportoletti P, Bernardi R, Bronson RT, Castillo-Martin M, Cordon-Cardo C, Lunardi A*, Pandolfi PP*. 2015. A genetic platform to model sarcomagenesis from primary adult mesenchymal stem cells. Cancer Discov. 2015 Jan 22. pii: CD-14-1022. PMID: 25614485
Liu XS, Chandramouly G, Rass E, Guan Y, Wang G, Hobbs RM, Rajendran A, Xie A, Shah JV, Davis AJ, Scully R, Lunardi A*, Pandolfi PP*. 2015. LRF maintains genome integrity by regulating the non-homologous end joining pathway of DNA repair. Nat Commun. Oct 8;6:8325. doi: 10.1038/ncomms9325. PMID: 26446488
Papa A, Wan L, Bonora M, Salmena L, Song MS, Hobbs RM, Lunardi A, Webster K, Ng C, Newton RH, Knoblauch N, Guarnerio J, Ito K, Turka LA, Beck AH, Pinton P, Bronson RT, Wei W, Pandolfi PP. 2014. Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell, 157(3):595-610. PMID:24766807
Wang G*, Lunardi A*, Zhang J, Chen Z, Ala U, Webster KA, Tay Y, Gonzalez-Billalabeitia E, Egia A, Shaffer DR, Carver B, Liu XS, Taulli R, Kuo WP, Nardella C, Signoretti S, Cordon-Cardo C, Gerald WL, Pandolfi PP. 2013. Zbtb7a suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat Genet, 45(7):739-46. PMID:23727861
Lunardi A, Ala U, Epping MT, Salmena L, Clohessy JG, Webster KA, Wang G, Mazzucchelli R, Bianconi M, Stack EC, Lis R, Patnaik A, Cantley LC, Bubley G, Cordon-Cardo C, Gerald WL, Montironi R, Signoretti S, Loda M, Nardella C, Pandolfi PP. 2013. A co-clinical approach identifies mechanisms and potential therapies for androgen deprivation resistance in prostate cancer. Nat Genet, 45(7):747-55. PMID:23727860
Reviews
Alaimo A, De Felice D, Genovesi S, Lorenzoni M, Lunardi A. 2021 Tune the channel: TRPM8 targeting in prostate cancer. Oncoscience. Sep 10;8:97-100. doi: 10.18632/oncoscience.543. eCollection 2021. PMID: 34514058
Rubert J, Schweiger PJ, Mattivi F, Tuohy K, Jensen KB, Lunardi A. 2020 Intestinal Organoids: A Tool for Modelling Diet-Microbiome-Host Interactions. Trends. Endocrinol Metab. Nov;31(11):848-858. doi: 10.1016/j.tem.2020.02.004. PMID: 33086077
Lunardi A, Pandolfi PP. 2014. A co-clinical platform to accelerate cancer treatment optimization. Trends Mol Med. 21(1):1-5. PMID:25466492
Lunardi A, Guarnerio J, Wang G, Maeda T, Pandolfi PP. 2013. Role of LRF/Pokemon in lineage fate decisions. Blood. 121(15):2845-53. PMID:23396304
Nardella C, Lunardi A, Patnaik A, Cantley LC, Pandolfi PP. 2011. The APL paradigm and the "co-clinical trial" project. Cancer Discov. 1(2):108-16. PMID:22116793
Collavin L, Lunardi A, Del Sal G. 2010. p53-family proteins and their regulators: hubs and spokes in tumor suppression. Cell Death Differ. Jun;17(6):901-11. PMID:20379196
Book chapters
Lunardi A, Nardella C, Clohessy JG, Pandolfi PP. 2014. Of model pets and cancer models: an introduction to mouse models of cancer. Cold Spring Harb Protoc. 1; (1):17-31. PMID:24371312
Links
https://pubmed.ncbi.nlm.nih.gov/?term=Lunardi%20Andrea&sort=date
https://lunardilab.wordpress.com
