Overview
Our group seeks to understand how cell division works in ovoid-shaped cocci and how this essential process is coordinated with cell elongation, using as a model Streptococcus pneumoniae.
S. pneumoniae (the pneumococcus) is an American-football shaped Gram-positive pathogenic coccus that lives in intimate association with its human host and is responsible for more than 1 million deaths per year worldwide. It colonizes asymptomatically the nasopharynx of healthy children and adults without apparent negative effects. However, it also causes a broad spectrum of diseases such as otitis media and sinusitis as well as pneumonia, bacteremia and meningitis. Pneumococcal diseases are prevalent among the most vulnerable members of society such as the infants, the elderly and other immunocompromised individuals. Penicillin and other β-lactam antibiotics have been successfully used for many years to treat pneumococcal infections. However, penicillin-resistant S. pneumoniae strains have increased dramatically since the 1980s and pose serious problems in the treatment of pneumococcal infections. Moreover, clinical isolates of S. pneumoniae resistant to other common antibiotics also continue to emerge, despite the existence of approved vaccines, calling for new lines of intervention. Since 2017, S. pneumoniae is included by the WHO as one of the superbugs in the list of the antibiotic-resistant bacteria for which research, discovery and development of new antibiotics is urgently needed. 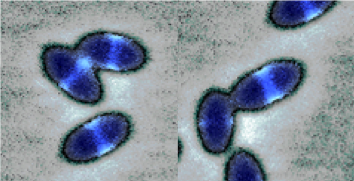
Research directions
The general goal of our research is to elucidate some important aspects of how ovoid-shaped Gram-positive cocci integrate the elongation and division processes, focusing on the essential proteins and their interacting partners to determine their structure and function
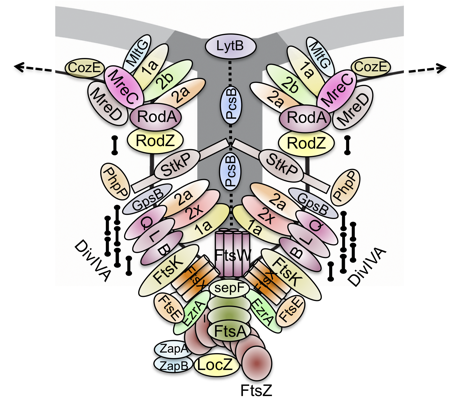 To achieve and maintain their cell shape during the cell cycle, bacteria use cytoskeletal proteins that, by polymerizing, translate molecular interactions into larger-scale cell dimensions. Most of the knowledge regarding the bacterial cell cycle comes from the well-studied rod-shaped models, Escherichia coli and Bacillus subtilis, that grow primarily by extension along their sidewall and then they divide, once each generation, by synthesizing a perpendicular division septum. Cell elongation is organized by MreB, an actin homolog that assembles the biosynthetic complex, known as the elongasome, which carries out peripheral cell wall synthesis, while cell division is organized by the tubulin homolog FtsZ which, in concert with another actin homolog, FtsA, assembles the biosynthetic complex, known as the divisome, which carries out septal cell wall synthesis. Similarly to the model rods, S. pneumoniae also grows by alternating sidewall synthesis and septal cell wall synthesis, albeit the latter prevails over the former, resulting in the characteristic ovoid-shape of pneumococci which are, for this reason, also referred to as ovococci. Consistent with this mode of growth, S. pneumoniae contains many of the components that constitute the elongasome and the divisome present in the model rods but, like the other cocci, lacks MreB. Moreover, instead of having two separate cell wall biosynthetic complexes, S. pneumoniae assembles a single complex at midcell that carries out both tasks. This complex requires a fine-tuned control, operated by the Eukaryotic-type Ser/Thr kinase, StkP, to integrate cell elongation with division, although the mechanism is not completely understood.
To achieve and maintain their cell shape during the cell cycle, bacteria use cytoskeletal proteins that, by polymerizing, translate molecular interactions into larger-scale cell dimensions. Most of the knowledge regarding the bacterial cell cycle comes from the well-studied rod-shaped models, Escherichia coli and Bacillus subtilis, that grow primarily by extension along their sidewall and then they divide, once each generation, by synthesizing a perpendicular division septum. Cell elongation is organized by MreB, an actin homolog that assembles the biosynthetic complex, known as the elongasome, which carries out peripheral cell wall synthesis, while cell division is organized by the tubulin homolog FtsZ which, in concert with another actin homolog, FtsA, assembles the biosynthetic complex, known as the divisome, which carries out septal cell wall synthesis. Similarly to the model rods, S. pneumoniae also grows by alternating sidewall synthesis and septal cell wall synthesis, albeit the latter prevails over the former, resulting in the characteristic ovoid-shape of pneumococci which are, for this reason, also referred to as ovococci. Consistent with this mode of growth, S. pneumoniae contains many of the components that constitute the elongasome and the divisome present in the model rods but, like the other cocci, lacks MreB. Moreover, instead of having two separate cell wall biosynthetic complexes, S. pneumoniae assembles a single complex at midcell that carries out both tasks. This complex requires a fine-tuned control, operated by the Eukaryotic-type Ser/Thr kinase, StkP, to integrate cell elongation with division, although the mechanism is not completely understood.
Most of our current work focus on the protein-protein interactions of the components that are crucial for streptococcal growth and division, their regulatory network and the dynamics of their cellular localization. Our aim is to identify the key elements and motifs in the selected proteins, and ultimately in the cell, that are indispensable to grow and divide.
This will also allow us to identify and validate novel targets that can be exploited to develope new antibiotics, for which there is an urgent need to counteract resistance.
Group members
- Orietta Massidda, PI
- Berenice Furlan, PhD student
- Sarah Costinas, PhD student
- Hamza Ur Rehman, Pre-doc
Collaborations
- Malcolm Winkler and Tiffany Tsui, Indiana University, Bloomington, US
- Pavel Branny and Linda Doubravová, Czech Academy of Sciences, Prague, CR
- Waldemar Vollmer, University of Newcastle, Newcastle Upon-Tyne, UK
- William Margolin, McGovern Medical School, Houston, US
- Jan-Willem Veening, University of Lausanne, CH
- Stefano Rossi, Università di Trento
Selected publications
Fulgenzio C, Massari M, Traversa G, Da Cas R, Ferrante G, Aschbacher R, Moser V, Pagani E, Vestri AR, Massidda O., Kurotschka PK. 2021. Impact of Prior Antibiotic Use in Primary Care on Escherichia coli Resistance to Third Generation Cephalosporins: A Case-Control Study. Antibiotics (Basel), 10(4):451.
Vollmer W, Massidda O., Tomasz A. 2019. The Cell Wall of Streptococcus pneumoniae. Microbiol Spectr 7(3). doi: 10.1128/microbiolspec.GPP3-0018-2018.
Cleverley RM, Rutter ZJ, Rismondo J, Corona F, Tsui HT, Alatawi FA, Daniel RA, Halbedel S, Massidda O., Winkler ME, Lewis RJ. 2019. The cell cycle regulator GpsB functions as cytosolic adaptor for multiple cell wall enzymes. Nat Commun, 10:261.
Zheng JJ, Perez AJ, Tsui HT, Massidda O., Winkler ME. 2017. Absence of the KhpA and KhpB (JAG/EloR) RNA-binding proteins suppresses the requirement for PBP2b by overproduction of FtsA in Streptococcus pneumoniae D39. Mol Microbiol, 106:793-814.
Rued BE, Zheng JJ, Mura A, Tsui HT, Boersma MJ, Mazny JL, Corona F, Perez AJ, Fadda D, Doubravová L, Buriánková K, Branny P, Massidda O., Winkler ME. 2017. Suppression and synthetic-lethal genetic relationships of ΔgpsB mutations indicate that GpsB mediates protein phosphorylation and penicillin-binding protein interactions in Streptococcus pneumoniae D39. Mol Microbiol, 103:931-957. Commentary in Lewis RJ. 2017. GpsB the truth is out there. Mol Microbiol, 103:913-918 (https://onlinelibrary.wiley.com/doi/full/10.1111/mmi.13612).
Mura A, Fadda D, Perez AJ, Danforth ML, Musu D, Rico AI, Krupka M, Denapaite D, Tsui HT, Winkler ME, Branny P, Vicente M, Margolin W*, Massidda O.. 2016. Roles of the Essential Protein FtsA in Cell Growth and Division in Streptococcus pneumoniae. J Bacteriol, 199:pii:e00608-16. (*co-last corresponding authors).
Holečková N, Doubravová L*, Massidda O.*, Molle V, Buriánková K, Benada O, Kofroňová O, Ulrych A, Branny P. 2014. LocZ is a new cell division protein involved in proper septum placement in Streptococcus pneumoniae. MBio, 6:e01700-14. (*co-corresponding authors).
Massidda O., Nováková, L, and W Vollmer. 2013. From models to pathogens: How much have we learned about Streptococcus pneumoniae cell division. Env Microbio,15: 3133-57.
Beilharz, K, Nováková, L, Fadda, D, Branny, P, Massidda O.*, Veening, JW*. 2012. Control of cell division in Streptococcus pneumoniae by the conserved Ser/Thr protein kinase StkP. Proc Natl Acad Sci, U.S.A.,109: E905–E913. (*co-last corresponding authors).
Palmieri C, Mingoia, M, Massidda O., Giovanetti E, Varaldo PE. 2012. Streptococcus pneumoniae transposon Tn1545/Tn6003 changes to Tn6002 due to spontaneous excision in circular form of the erm(B)- and aphA3-containing macrolide-aminoglycoside-streptothricin (MAS) element. Antimicrob Agents Chemother, 56:5994–5997.
Maggi, S, Massidda O., Luzi, G, Fadda, D, Paolozzi, L, Ghelardini, P. 2008. Division protein interaction web: identification of a phylogenetically conserved common interactome between Streptococcus pneumoniae and Escherichia coli. Microbiology (SGM), 154:3042–3052. Le Gouëllec, A, Roux, L, Fadda, D, Massidda, O, Vernet, T, Zapun, A. 2008. Roles of pneumococcal DivIB in cell division. J Bacteriol, 190:4501–4511.
Fadda, D, Santona, A, D’Ulisse, V, Ghelardini, P, Ennas, MG, Whalen, MB, Massidda O.. 2007. Streptococcus pneumoniae DivIVA: localization and interactions in a MinCD-free context. J Bacteriol, 189:1288–1298. Journal Highlight in "How DivIVA Controls Morphology During Cell Division in S. pneumoniae". Microbe vol 2, number 3, 2007 (https://iris.unitn.it/retrieve/handle/11572/187531/157696/Fadda%20et%20al_Journal%20Highlight.pdf); Guest Commentary: Vicente M & García-Ovalle M,. 2007. Making a point: the role of DivIVA in streptococcal polar anatomy”. J Bacteriol, 89:1185-8 (https://jb.asm.org/content/189/4/1185).
Vicente, M, Hodgson, J, Massidda O., Tonjum, T, Henriques-Normark, B, and Ron, EZ. 2006. The fallacies of hope: will we discover new antibiotics to combat pathogenic bacteria in time? FEMS Microbiol Rev, 30: 841–852.
Smith AM, Feldman, C, Massidda O., McCarthy, K, Ndiweni, D, Klugman, KP. 2005. Altered PBP 2A and its role in the development of penicillin, cefotaxime, and ceftriaxone resistance in a clinical isolate of Streptococcus pneumoniae. Antimicrob Agents Chemother, 49:2002–2007.
Lara B, Rico AI, Petruzzelli S, Santona A, Dumas J, Biton J, Vicente M, Mingorance J, Massidda O.. 2005. Cell division in cocci: localization and properties of the Streptococcus pneumoniae FtsA protein. Mol Microbiol, 55:699–711.
Fadda D, Pischedda C, Caldara F, Whalen MB, Anderluzzi D, Domenici E, Massidda O.. 2003. Characterization of divIVA and other genes located in the chromosomal region downstream of the dcw cluster in Streptococcus pneumoniae. J Bacteriol, 185:6209–6214.
Carettoni D, Gómez-Puertas P, Yim, L, Mingorance J, Massidda O., Vicente M, Valencia A, Domenici E, Anderluzzi D. 2003. Phage-display and correlated mutations identify an essential region of subdomain 1C involved in homodimerization of Escherichia coli FtsA. Proteins, 50:192–206.
Massidda O., Anderluzzi D, Friedli L, Feger G 1998. Unconventional organization of the division and cell wall gene cluster of Streptococcus pneumoniae. Microbiology (SGM), 144:3069–3078.
