Overview
The survival and replication of viruses depends on their ability to control, hijack, and profit from the activity of the host cell. Deciphering the interaction between the virus and the host cell throughout its life cycle is instrumental not only for the identification of novel potential opportunities for antiviral intervention but also for a deeper mechanistic knowledge of the cellular machinery. We are interested in both aspects: exploring the way viruses interact with host cellular components and improving our understanding of the host pathways involved. 
Research directions
Virus replication is regulated by the complex and not always well understood activity of multifunctional viral proteins to gain optimal advantage of the host machinery. On the other hand, as a result of its co-evolution with viral parasites, the host cell has developed mechanisms to interfere with the viral replication. We are exploring both aspects of this host-parasite relationship.
-
SERINC proteins
After a long quest to identify the mechanism by which Nef potentiates the infectivity of HIV, we have identified the cellular protein SERINC5 as an inhibitor of retroviruses (Rosa et al., Nature, 526, 2015). SERINC proteins are multipass transmembrane proteins highly conserved throughout eukaryotes. We found that SERINC5 is incorporated into viruses and interferes with the virus particles ability to enter target cells. Our studies established that Nef of primate lentiviruses (HIV-1, HIV-2 and SIV), as well as other retrovirus factors from gammaretroviruses, glycoGag (Pizzato M., " PNAS 107, 2010 ), and equine infectious anemia virus, S2 (Chande et al., PNAS, 113, 2016); counteract SERINC5 by promoting its downregulation to the late endosomal compartment. The emergence of a SERINC antagonizing activity in different groups of retroviruses provide a compelling example of convergent evolution and underscores the important antiviral role of SERINC5.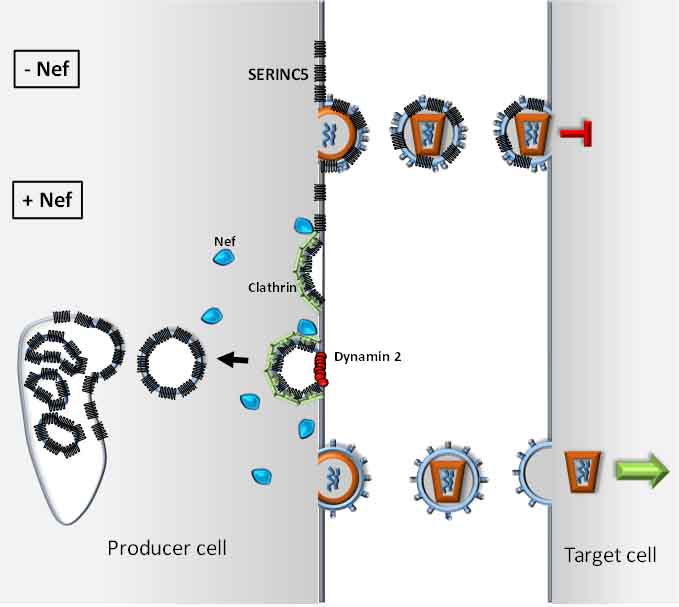
We are now investigating not only the antiviral activity of SERINC proteins, but also its broader functions in cell biology and in immunity, which remain mostly unknown.
Group members
- Massimo Pizzato, PI
- Cinzia Bertelli, PhD student
- Caterina Bianchi, MSc student
Collaborations
- Peter Cherepanov, Crick Institute, London
- Jeremy Luban, University of Massachusetts Medical School, USA
- Felipe-Diaz Griffero, Albert Einstein College of Medicine
- Yasuhiro Takeuchi, UCL, London
- Oliver Fackler, University Hospital Heidelberg, Germany
Funding
Bando: PRIN 2022 (D.D. 104/22)
The cellular function of SERINC proteins and their role in retroviral infection
Massimo Pizzato, Coordinatore
Codice Protocollo: 2022RYNKB5 CUP: E53D23010040006

Selected publications
Firrito C, Bertelli C, Vanzo T, Chande A, Pizzato M. “SERINC5 as a New Restriction Factor for Human Immunodeficiency Virus and Murine Leukemia Virus.” Annu Rev Virol. 29;5(1):323-340 (2018).
Sood C., Marin M., Chande A., Pizzato M. and Melikyan G.B. “SERINC5 Inhibits HIV-1 Fusion Pore Formation by Promoting Functional Inactivation of Envelope Glycoproteins”. J. Biol. Chem. jbc.M117.777714. (2017).
Chande A, Cuccurullo EC, Rosa A, Ziglio S, Carpenter S, Pizzato M. “S2 from Equine infectious anemia virus is an infectivity factor which counteracts the retroviral inhibitors SERINC5 and SERINC3”. PNAS, 113:13197-13202 (2016).
Trautz B, Pierini V, Wombacher R, Stolp B, Chase AJ, Pizzato M, Fackler OT. “Antagonism of the SERINC5 Particle Infectivity Restriction by HIV-1 Nef Involves Counteraction of Virion-associated Pools of the Restriction Factor.” Journal of virology. JVI.01246-16 (2016)
Heigele A, Kmiec D, Regensburger K, Langer S, Peiffer L, Stürzel CM, Sauter D, Peeters M, Pizzato M, Learn GH, Hahn B, Kirchhoff F. “The Potency of Nef-mediated SERINC5 Antagonism Correlates with the Prevalence of Primate Lentiviruses in the Wild”, Cell Host and Microbe, 20: 381-391 (2016).
Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M. “HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation”. Nature, 526: 121-7. (2015).
Pizzato M, McCauley SM, Neagu MR, Pertel T, Firrito C, Ziglio S, Dauphin A, Zufferey M, Berthoux L & Luban J. "Lv4 Is a Capsid-Specific Antiviral Activity in Human Blood Cells That Restricts Viruses of the SIVMAC/SIVSM/HIV-2 Lineage Prior to Integration". PLoS pathogens 11, e1005050 (2015).
Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, Bisiaux A, Albert ML, Strambio-De-Castillia C, Mothes W, Pizzato M, Grutter MG & Luban J. "TRIM5 is an innate immune sensor for the retrovirus capsid lattice". Nature 472, 361-365 (2011).
Lai RP, Yan J, Heeney J, McClure MO, Gottlinger H, Luban J & Pizzato M. "Nef decreases HIV-1 sensitivity to neutralizing antibodies that target the membrane-proximal external region of TMgp41". PLoS pathogens 7, e1002442 (2011).
Pizzato M. "MLV glycosylated-Gag is an infectivity factor that rescues Nef-deficient HIV-1". PNAS 107, 9364-9369 (2010).
Pizzato M, Erlwein O, Bonsall D, Kaye S, Muir D & McClure MO. "A one-step SYBR Green I-based product-enhanced reverse transcriptase assay for the quantitation of retroviruses in cell culture supernatants". Journal of virological methods 156, 1-7 (2009).
Takeuchi Y, McClure MO & Pizzato M. "Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research". Journal of virology 82, 12585-12588 (2008).
